Semantic types
Unlike Excel, table columns in Datagrok are strongly-typed, meaning all cells are of one of the predefined data
types (string, int, bigint, qnum, double, datetime, bool). In addition to that, a column might also have
semantic type associated with it. Semantic type identifies the meaning of the data. For instance, a column could be of
the data type "string", but the semantic type would be "country".
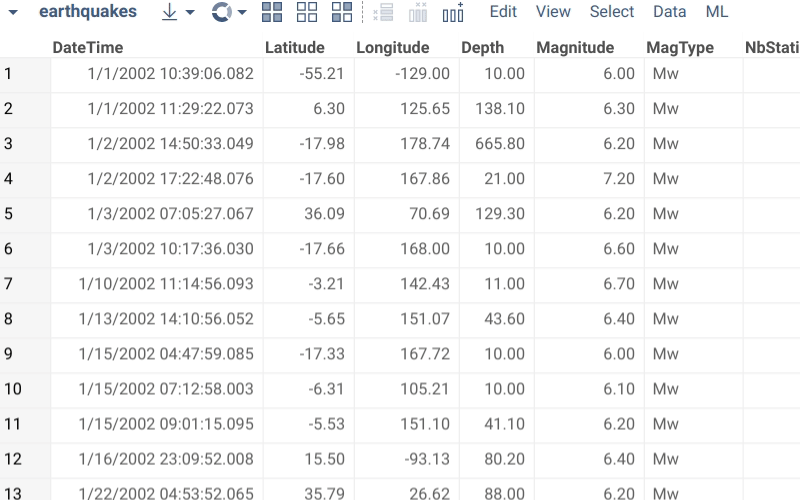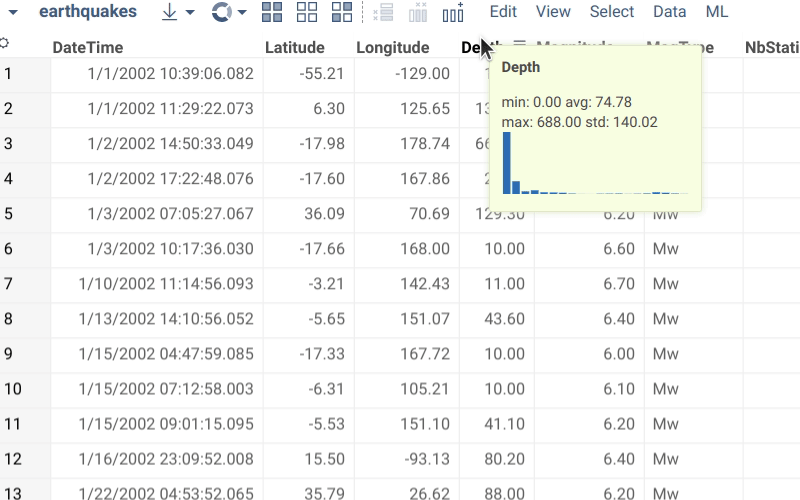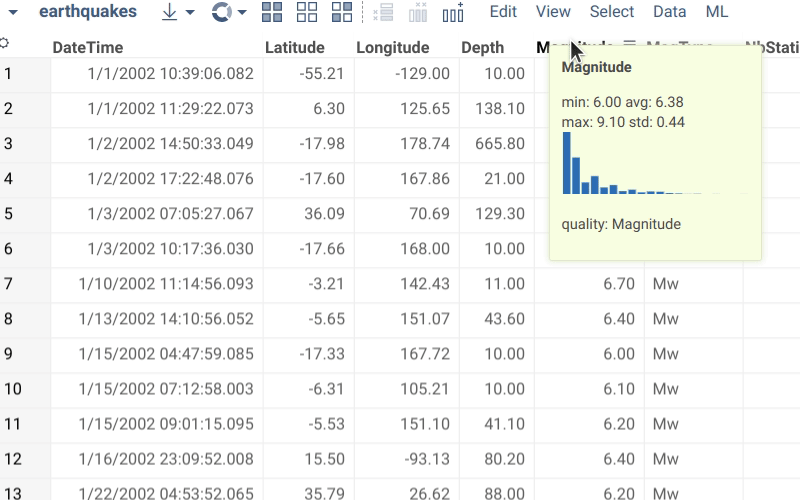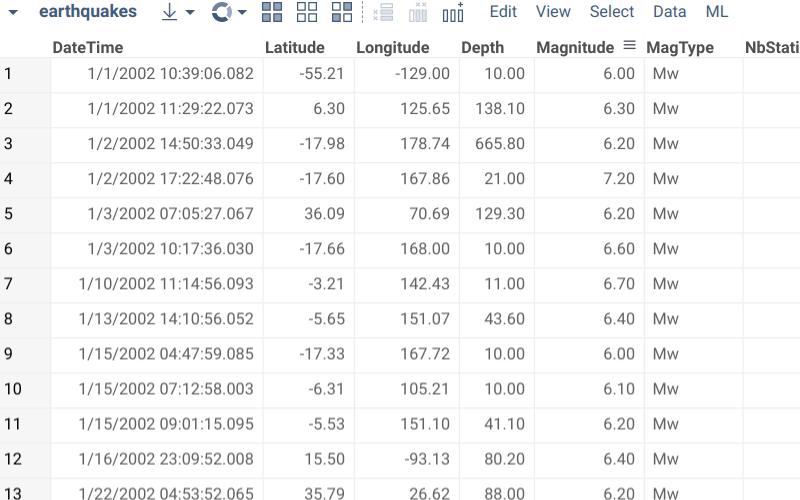
Semantic types are used in several ways:
- Rendering. It is possible to define a custom renderer defined for the semantic type. For instance, in
the
#{x.Demo:Smiles}project strings with the semantic type "molecule" get rendered as chemical structures. - Viewers. Some viewers, such as Map, use semantic type to determine whether a viewer can be visualized against specific datasets, as well as for the initial choice of columns to visualize.
- Functions. Function parameters could be annotated with the semantic type. This is used for automatic suggestions of applicable functions.
- Predictive models. Just like functions, semantic type annotations are used to determine whether or not a model is applicable to the dataset.
A column can have only one semantic type. It is stored in the column tags and can be either detected automatically by semantic type detectors or set manually.
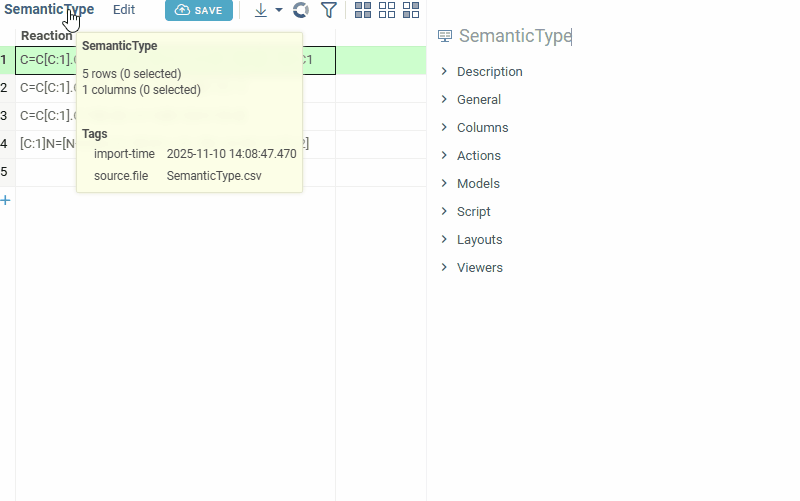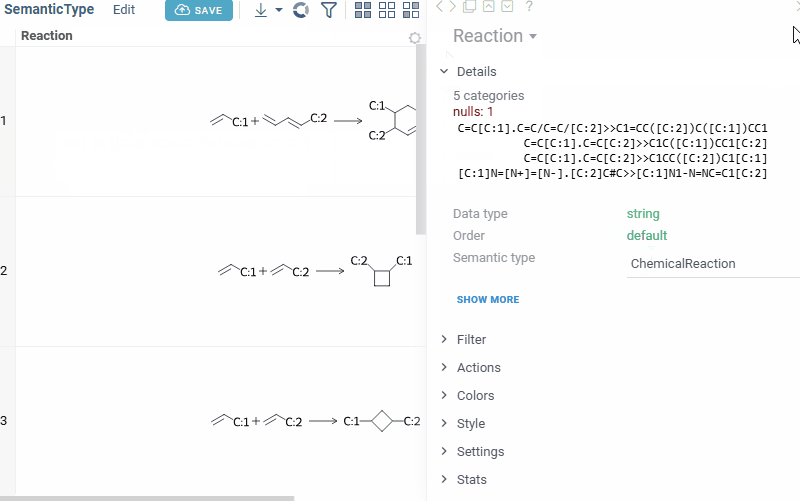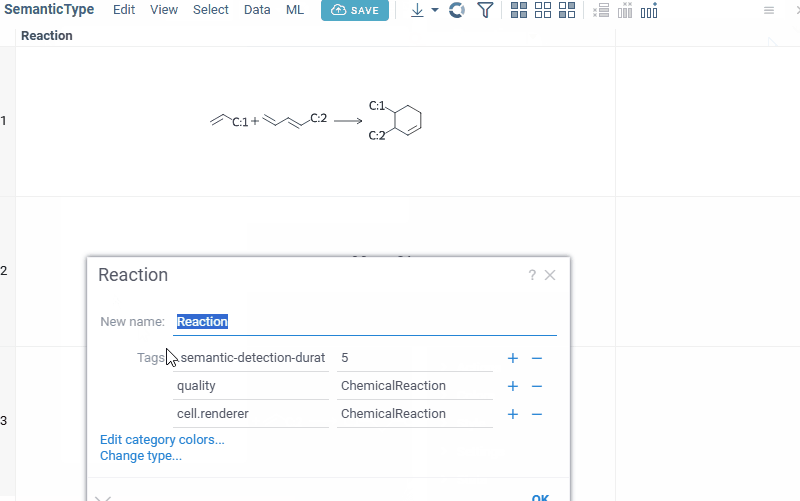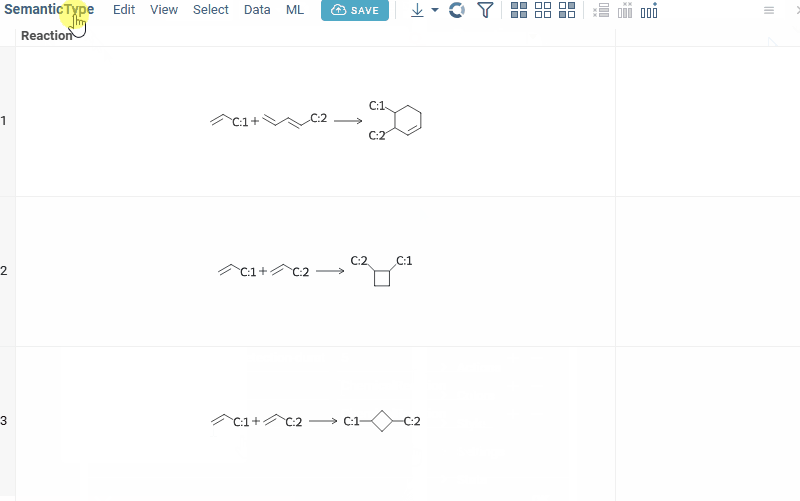
Manually setting semantic types
Open the Context Panel and go to the Details tab to set the 'Semantic type' field.
Alternatively, open column properties (click a column and press F2, or right-click the column and select Column Properties...) and set the quality and cell.renderer tags.
Automatic semantic type detection
Out of the box, the following semantic types are automatically detected based on the content:
| Semantic type | Value | Required plugins |
|---|---|---|
| Cheminformatics types | ||
| Molecule | Molecule | Chem |
| Substructure | Substructure | Chem |
| Chemical mixture (Mixfile format) | ChemicalMixture | Chem |
| Bioinformatics types | ||
| Macromolecule | Macromolecule | Bio |
| HELM | HELM | Bio |
| Phylogenetic trees | Newick | Bio |
| Molecule 3D | Molecule3D | Biostructure Viewer |
| PDB identifiers | PDB_ID | Biostructure Viewer |
| General types | ||
| Text | Text | - |
| Duration | Duration | - |
| Gender | Gender | - |
| Money | Money | - |
| Image | Image | - |
| File | File | - |
| URL | URL | - |
| IP address | IP Address | - |
| Contact information | ||
Email Address | - | |
| Phone number | Phone Number | - |
| Geographic data | ||
| City | City | - |
| Country | Country | - |
| State | State | - |
| County | County | - |
| Place name | Place Name | - |
| Zip code | Zip Code | - |
| Area code | Area Code | - |
| Street address | Street Address | - |
| Latitude | Latitude | - |
| Longitude | Longitude | - |
| Specialized types | ||
| Curve data | fit | Curves |
| PNG images | rawPng | Power Grid |
Custom semantic types
Datagrok has been designed to be as extensible as possible, so naturally it is possible to define your own custom semantic types, along with the type auto-detection functions, renderers, comparers, and so on. To do that, check out this article .
See also: