Chemprop
Property prediction plays a key role in drug discovery and materials science, helping researchers estimate characteristics like toxicity, bioactivity, and solubility.
We have now integrated Chemprop, a PyTorch-based framework, into our platform, enabling easy access with just a few clicks.
Model parameters
| Parameter | Description | Default |
|---|---|---|
| dataset_type | Type of dataset (e.g., classification or regression). Determines the training loss function. | Regression |
| metric | Evaluation metric. Does not impact training loss. Defaults to "AUC" for classification, "RMSE" for regression. | None |
| multiclass_num_classes | Number of classes when running multiclass classification. | 3 |
| num_folds | Number of folds when performing cross validation. | 1 |
| data_seed | Seed for data splitting. For multiple folds, increments by 1 for each fold. | 0 |
| split_sizes | Proportions for train/validation/test splits. | 0.8, 0.1, 0.1 |
| split_type | Data splitting method (e.g., random, cross-validation). | Random |
| activation | Activation function used in the model (e.g., ReLU, tanh). | ReLU |
| atom_messages | Uses messages on atoms rather than bonds. | False |
| message_bias | Adds bias to linear layers. | False |
| ensemble_size | Number of models in the ensemble. | 1 |
| message_hidden_dim | Hidden layer dimensionality in the message-passing network. | 300 |
| depth | Number of message-passing steps. | 3 |
| dropout | Dropout probability for training. | 0.0 |
| undirected | Sums bond vectors to treat edges as undirected. | False |
| ffn_hidden_dim | Hidden dimension size for the feed-forward network. | 300 |
| ffn_num_layers | Number of layers in the feed-forward network after message-passing encoding. | 2 |
| epochs | Total number of training epochs. | 50 |
| batch_size | Batch size for training. | 64 |
| warmup_epochs | Linear increase of learning rate from initial to max, then exponential decay to final. | 2.0 |
| init_lr | Starting learning rate for training. | 0.0001 |
| max_lr | Peak learning rate for training. | 0.001 |
| final_lr | Ending learning rate after decay. | 0.0001 |
| no_descriptor_scaling | Disables feature scaling for descriptors. | False |
Train
To train a model with Chemprop:
- Prepare dataset
- Ensure it includes a column with SMILES or molblocks.
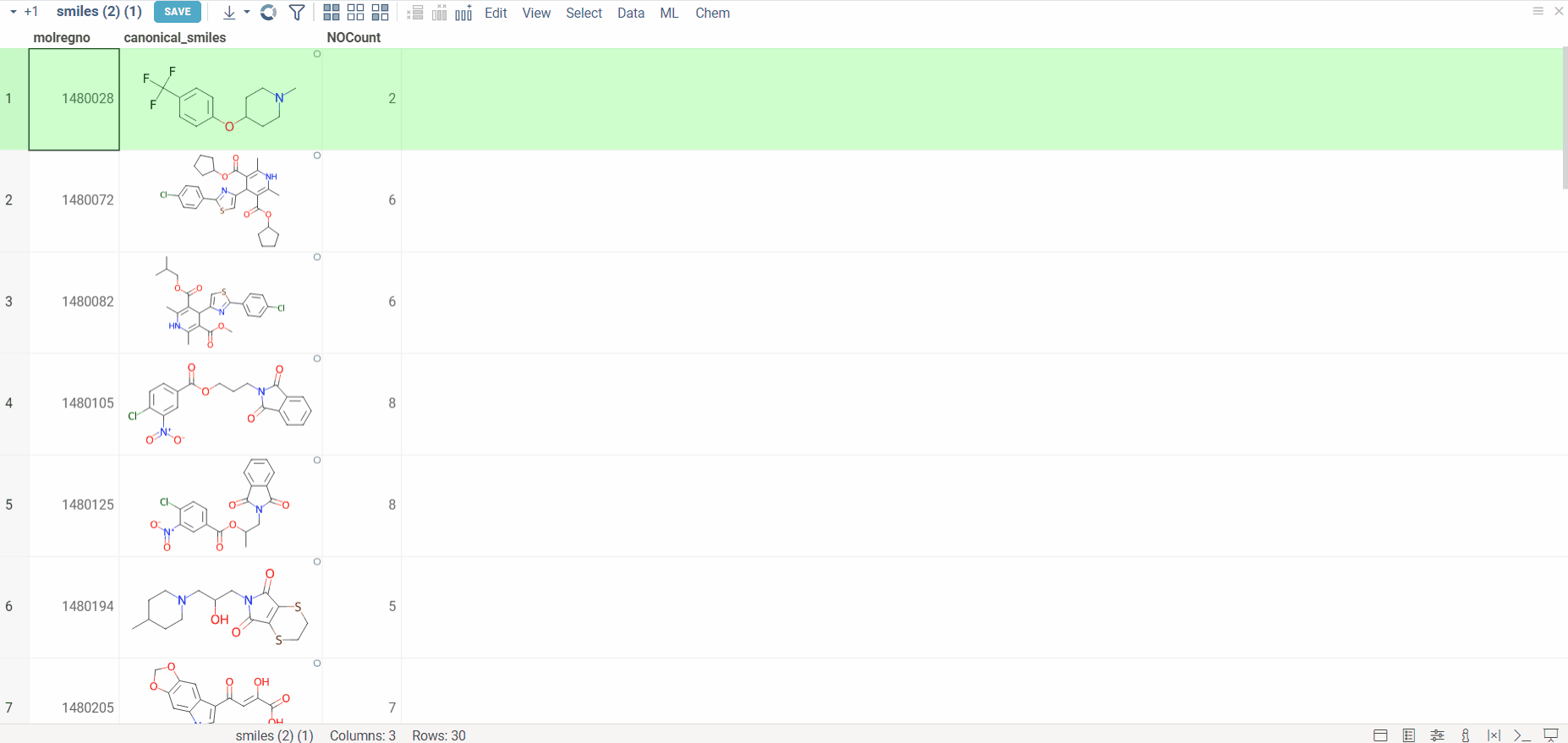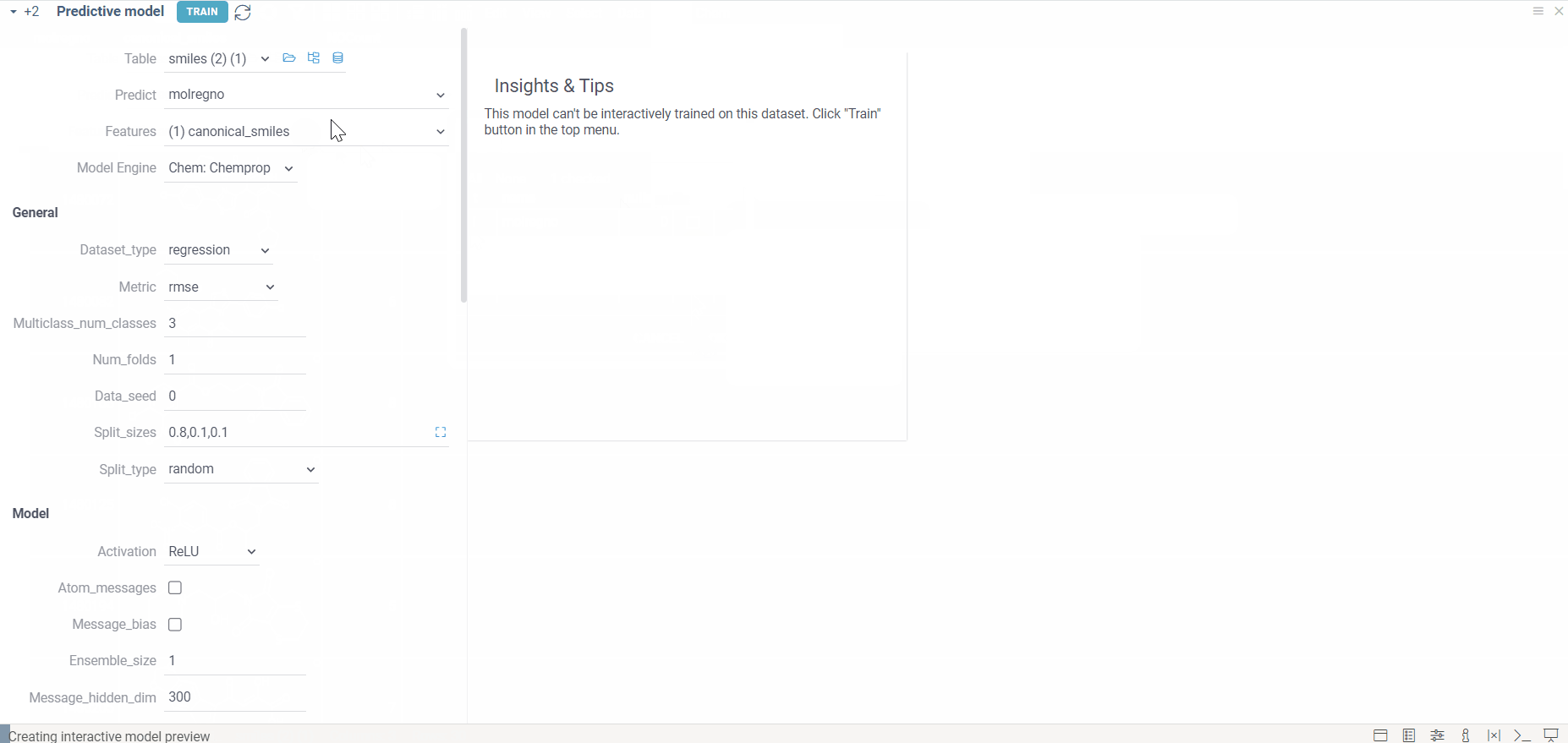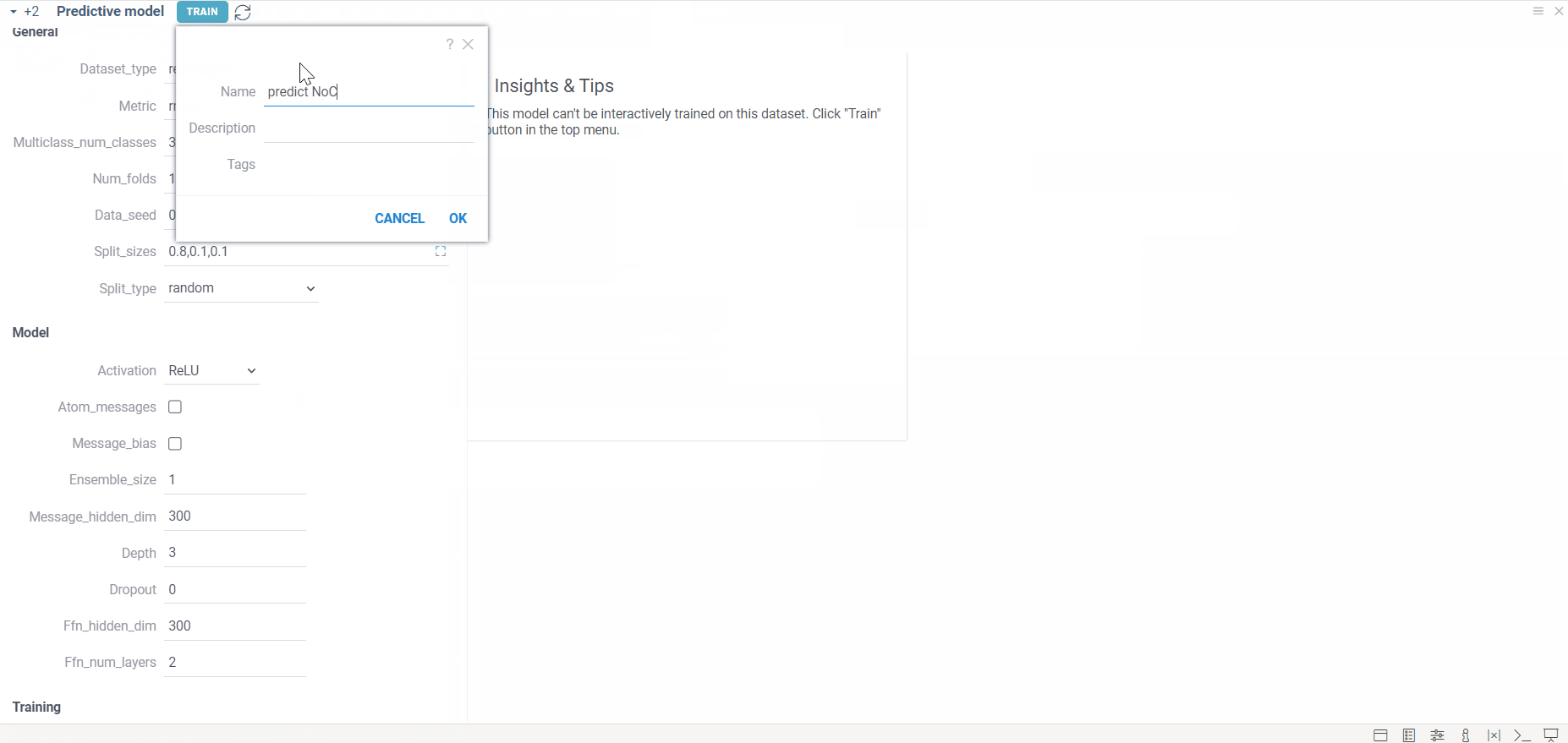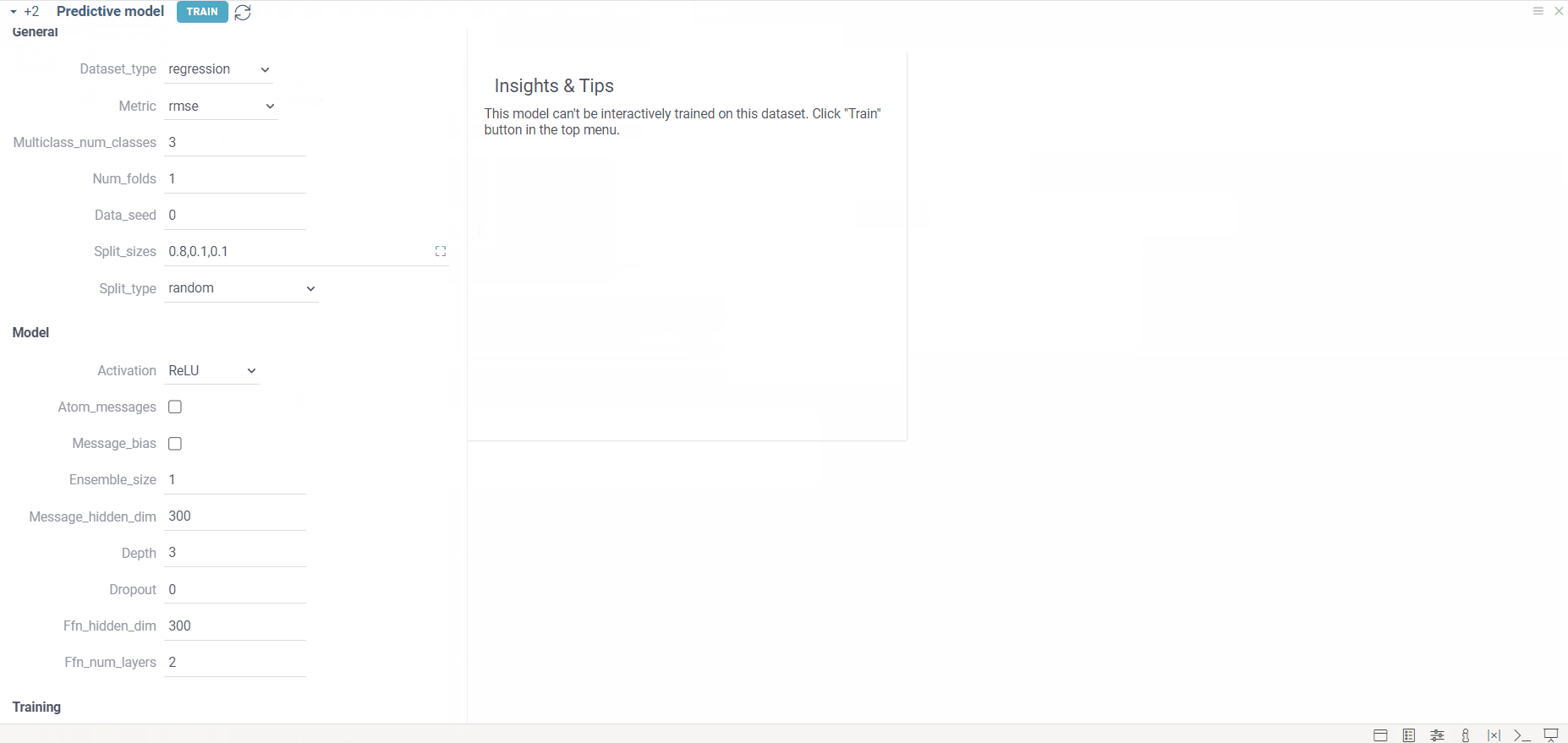
- Set up the model
- Go to ML > Models > Train Model….
- Select dataset, specify the target column (values to predict), and the feature column (SMILES or molblocks).
- Choose Chemprop as the model engine
- From available engines, select Chemprop.
- Adjust model settings
- Configure training parameters (epochs, batch size, learning rate) and evaluation metrics as needed.
- Train model
- Click Train to start the training process and generate the model.
Predict
Once training is complete, follow these steps to apply a trained model for predictions:
- Select the dataset
- Choose the table containing the features for prediction.
- Apply the model
- Go to ML > Models > Apply Model.
- Select the trained model you wish to use from the available options.
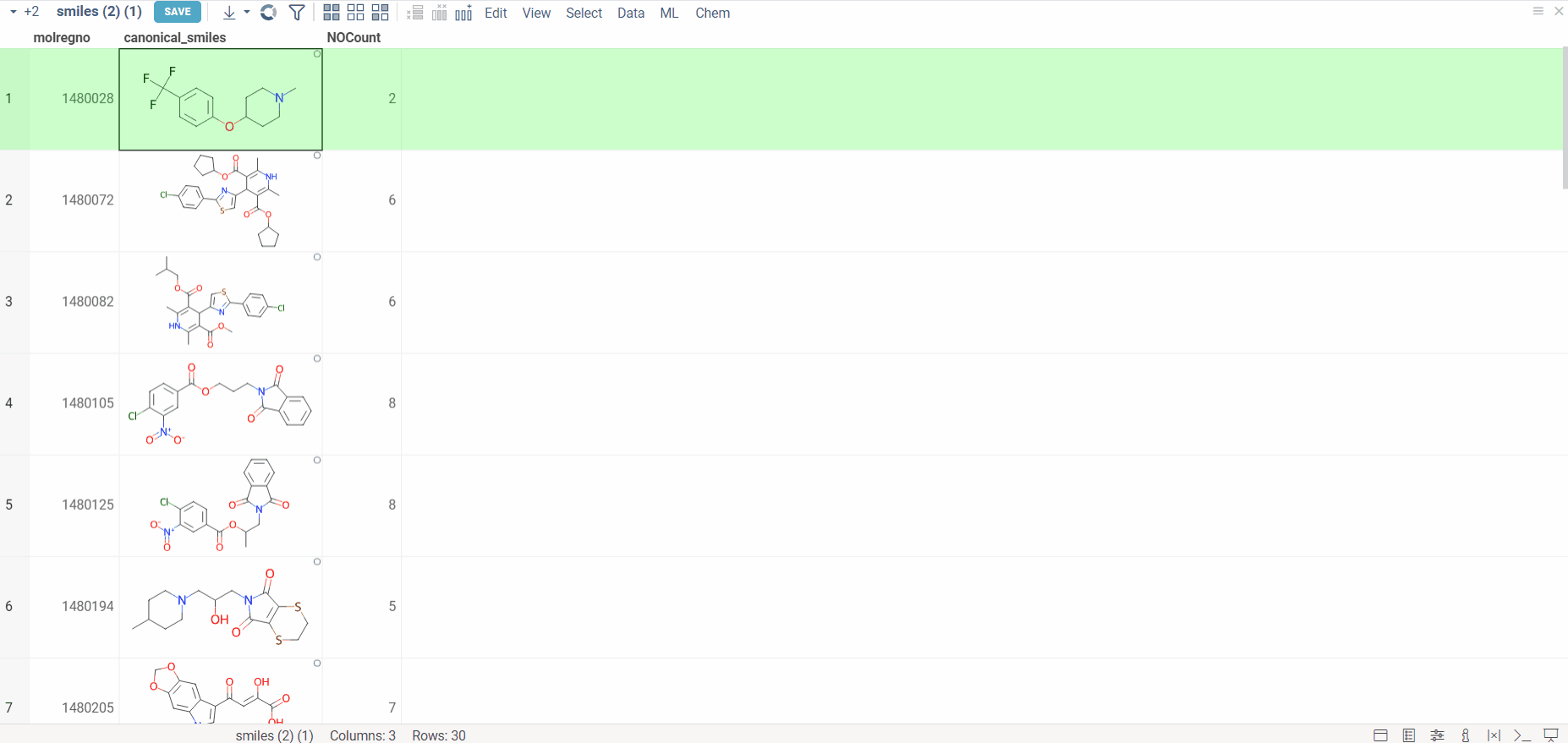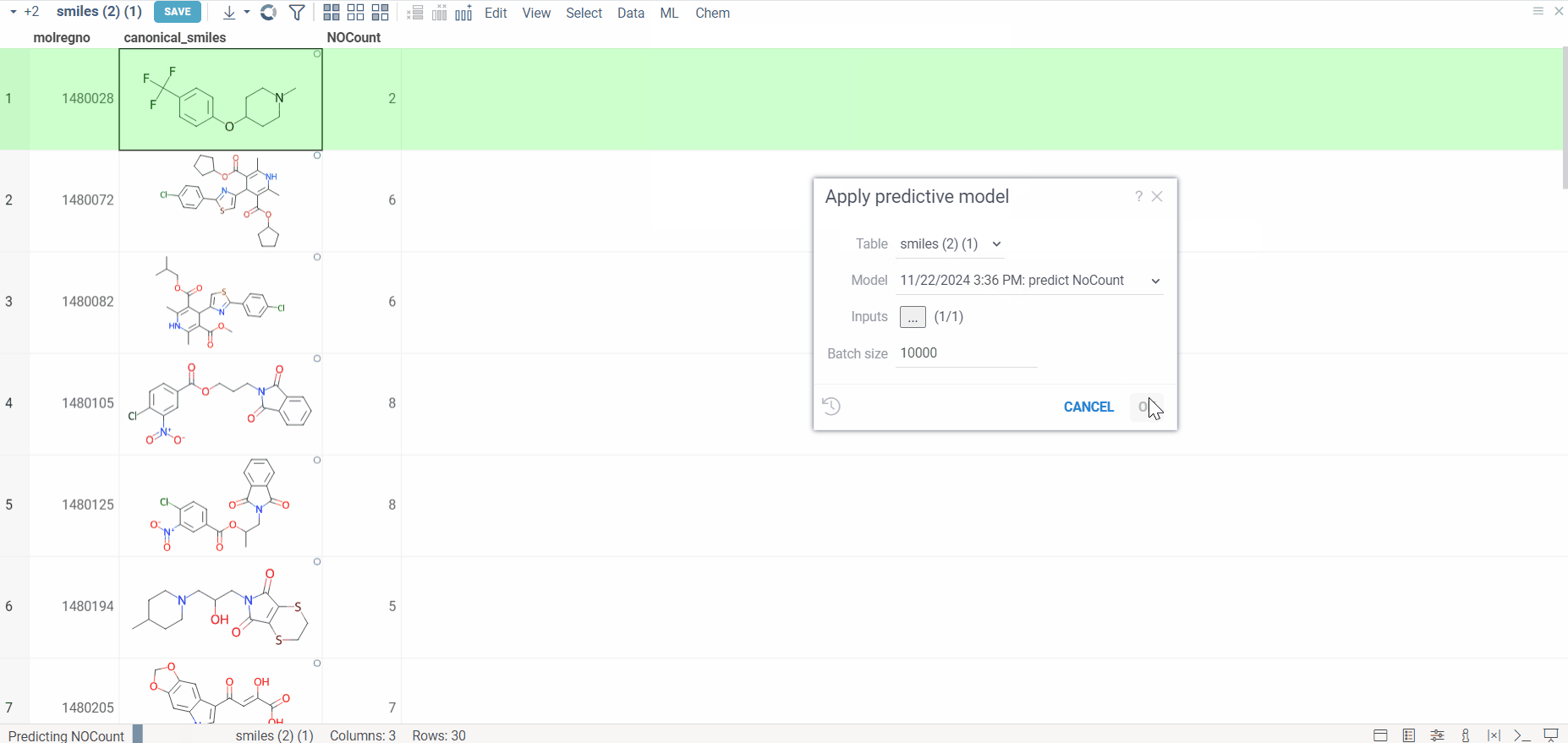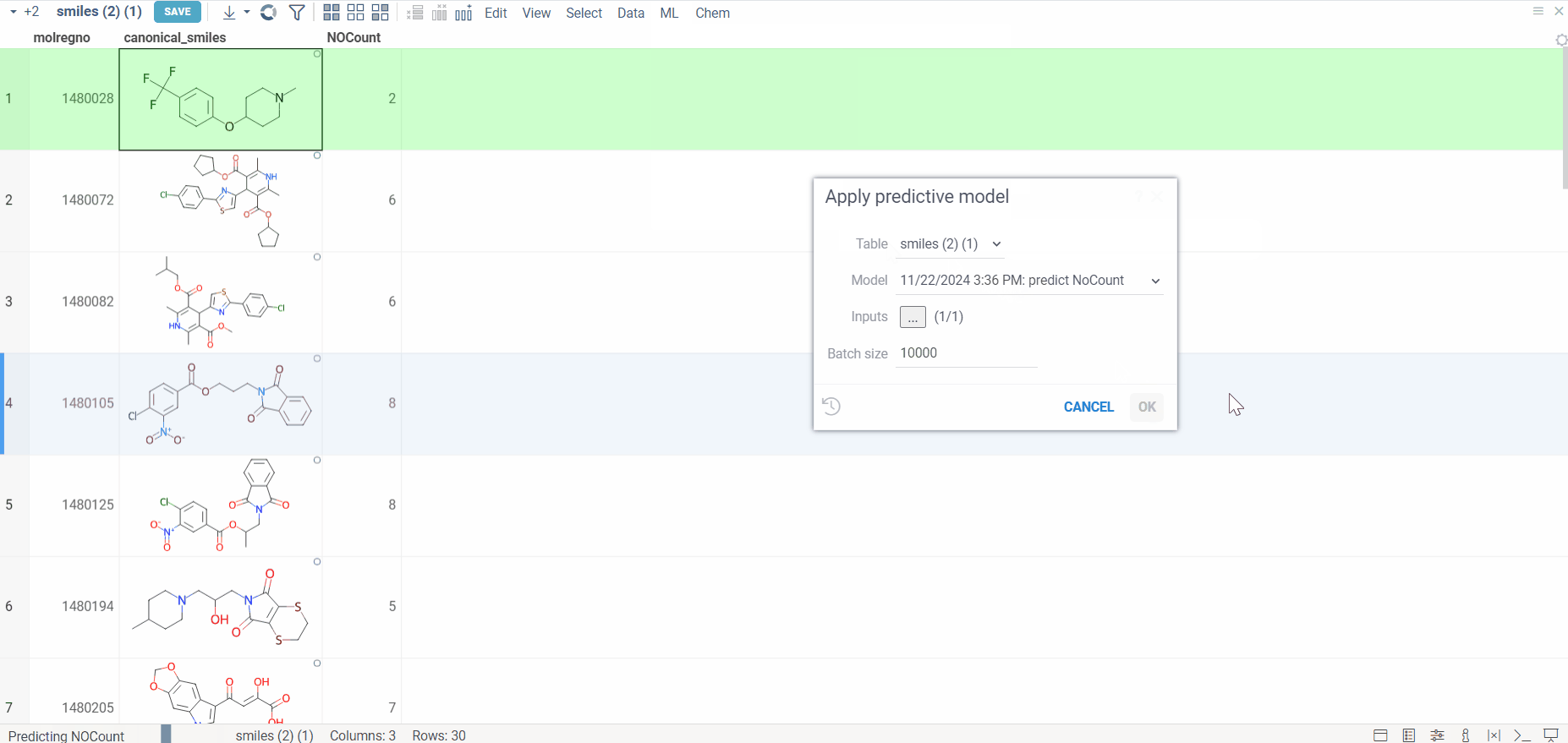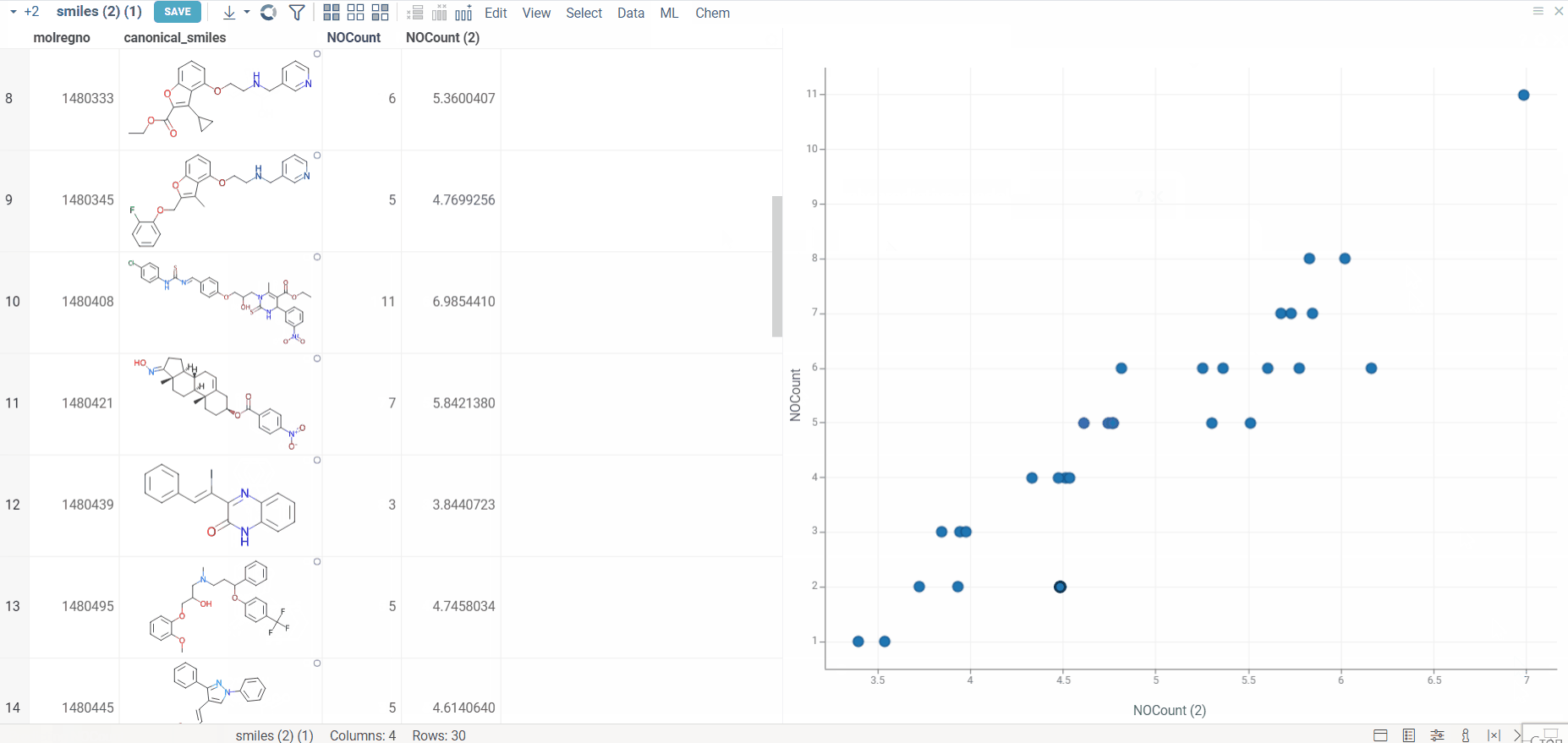
- Generate predictions
- Click Apply Model to make predictions on the selected dataset.