Bioinformatics
Requirements
To access the bioinformatics functionality, install these packages using the Package Manager (on the Sidebar, click Manage > Packages):
- Required. Bio.
- Optional. Biostructure Viewer: Visualization of macromolecules in 3D.
- Optional. Helm: Rendering, editing, managing monomer libraries.
- Optional. Peptides: SAR analysis for sequences.
Datagrok lets you work with macromolecules both on the macro (sequence) level and atomic level:
- Data visualization and exploration
- Support for multiple formats, such as FASTA (DNA/RNA/protein), delimiter-separated FASTA, HELM, PDB, and others. Handles nucleotides, natural and non-natural peptides, 3D-structures, and other modalities.
- Automatic detection of sequences upon data import.
- Flexible and fast spreadsheet that shows both macro and small molecules.
- Interactive visualization of biological data.
- Customizable info panes with information about macromolecules and context actions.
- Sequence editing, search, and filtering.
- Sequence analysis
- Structure-Activity Relation (SAR) analysis
- Activity cliffs
- A fit-for-purpose application for SAR analysis for peptides
- A comprehensive ML toolkit for clustering, dimensionality reduction techniques, imputation, PCA/PLS, and other tasks. Built-in statistics.
- Flexible reporting and sharing options, including dynamic dashboards.
- Oligonucleotides chemical modifications and format conversion.
- Connection to chemistry level: split to monomers, and get the atomic-level structure.
- Extensible environment
- Ability to add or customize any functionality using scripts in Python, R, Matlab, and other supported languages.
- Ability to create custom plugins and fit-for-purpose applications.
- Enterprise-grade platform for efficient data access and management.
Data access
Datagrok provides a single, unified access point for organizations. You can connect to local file storage, clouds (Amazon S3, Google cloud, etc), SQL and NoSQL databases or any other of the 30+ supported data sources, retrieve data, and securely share data with others. Datagrok can ingest data in multiple file formats (such as Fasta or CSV) and multiple notations for nucleotide and amino acid sequences, with natural and modified monomers, aligned and non-aligned forms.
You can also create macromolecule queries against data sources using built-in querying tools. To learn more about querying data and data access in general, see the Access section of our documentation.
Exploring biological data
Datagrok provides a range of tools for analyzing macromolecules (Top Menu > Bio). In addition, Datagrok provides a comprehensive machine learning toolkit for clustering, dimensionality reduction techniques, imputation, PCA/PLS, and other tasks (Top Menu > ML). Some of these tools can be applied directly to macromolecules.
When you open a dataset, Datagrok automatically detects macromolecules and makes available macromolecule-specific context actions. For example, when you open a CSV file containing macromolecules in the FASTA format, the following happens:
- Data is parsed, and the semantic type macromolecule is assigned to the corresponding column.
- Macromolecules are automatically rendered in the spreadsheet.
- Column tooltip now shows the sequence composition.
- Default column filter is now a subsequence search.
- A top menu item labeled Bio appears.
- Column and cell properties now show macromolecule-specific actions, such as sequence renderer and libraries options, sequence preview, and macromolecule space preview.
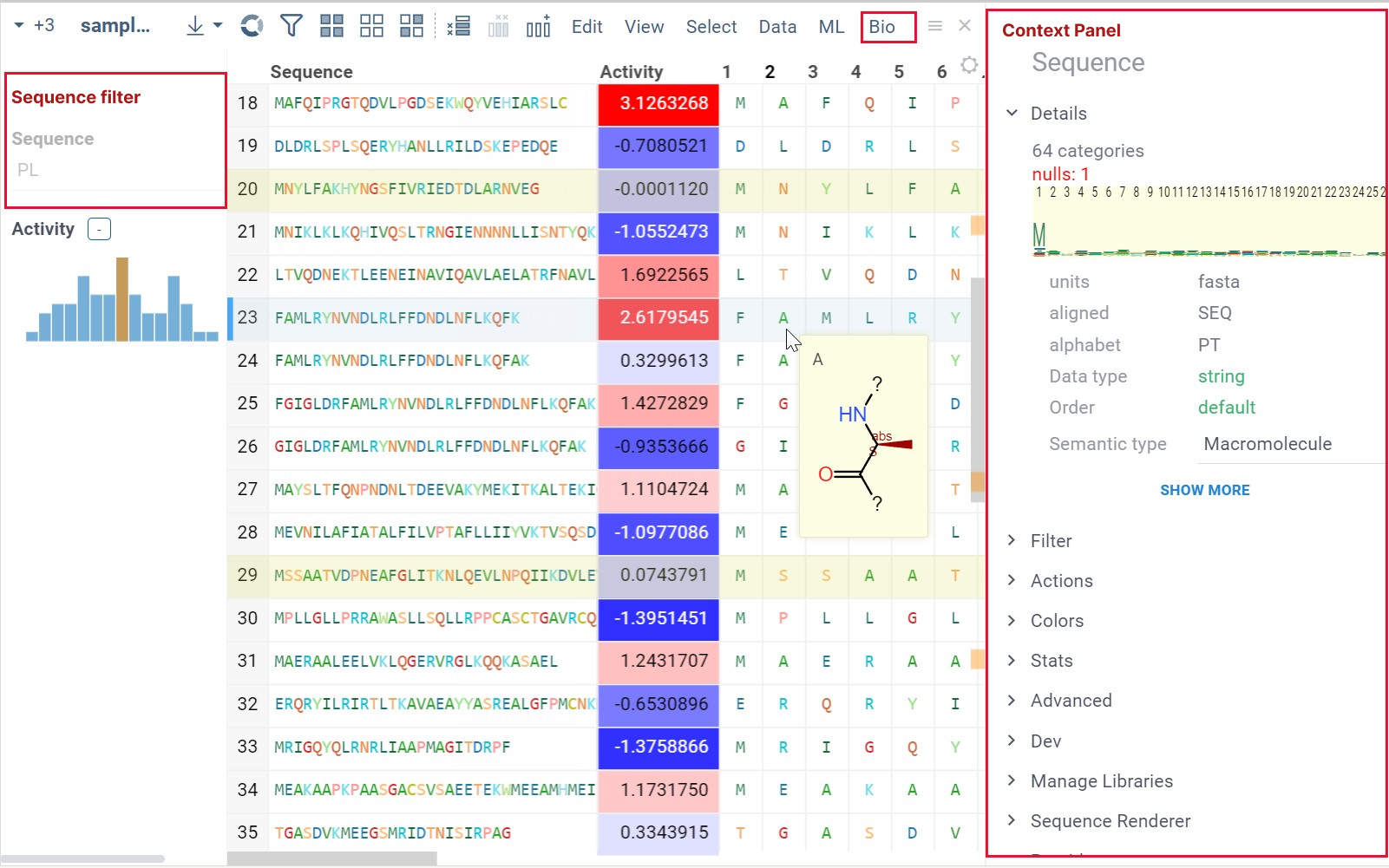
When you click on a column with macromolecules, you see the following in the Context Panel:
- Filter
- Manage Libraries
- Sequence Renderer: Rendering options.
- Peptides: From this pane, you can launch the SAR analysis for peptides.
Info pane options
Some info panes can be customized. To reveal an info pane's available options, hover over it:
- View and/or edit the underlying script (click the Script icon).
- Change parameters (click the Parameter icon).
- Change the info pane's settings (click the Gear icon).
- Append info pane as a column (click the More actions icon and select Add as a column).
To learn how to customize and extend the platform programmatically, see the Develop section of our documentation.
Info panes be extended with functions in any supported language.
Spreadsheet
The spreadsheet lets you visualize and edit sequences and macromolecules. You can add new columns with calculated values, interactively filter and search rows, color-code columns, pin rows or columns, set edit permissions, and more.
- HELM
- FASTA
- PDB
HELM is used for macromolecules with non-natural monomers, circular and branching structures. The structures are displayed with colors corresponding to each monomer.
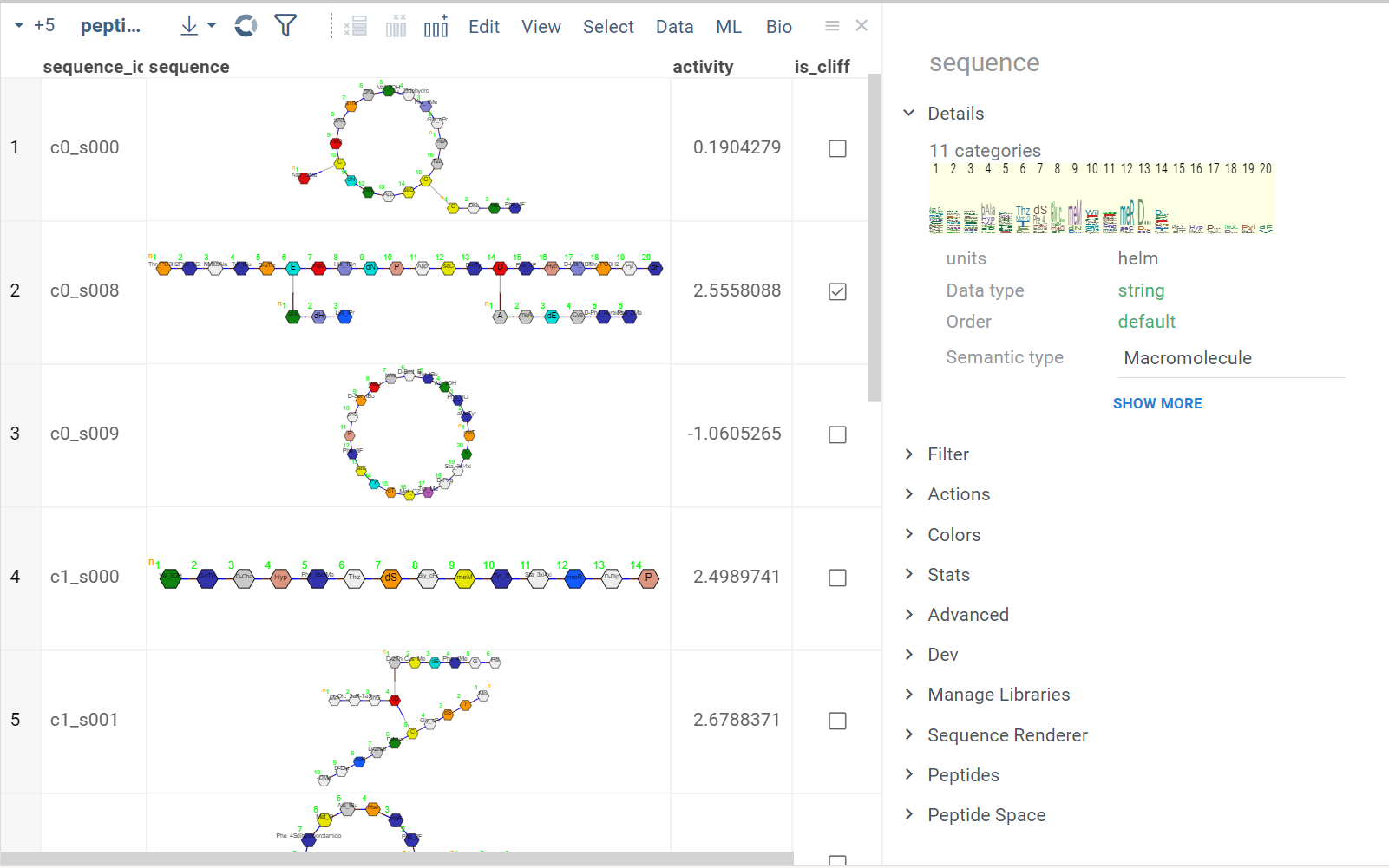
Peptide sequences are color-coded based on amino acid properties. DNA sequences are colored-coded to distinguish between various nucleotides.
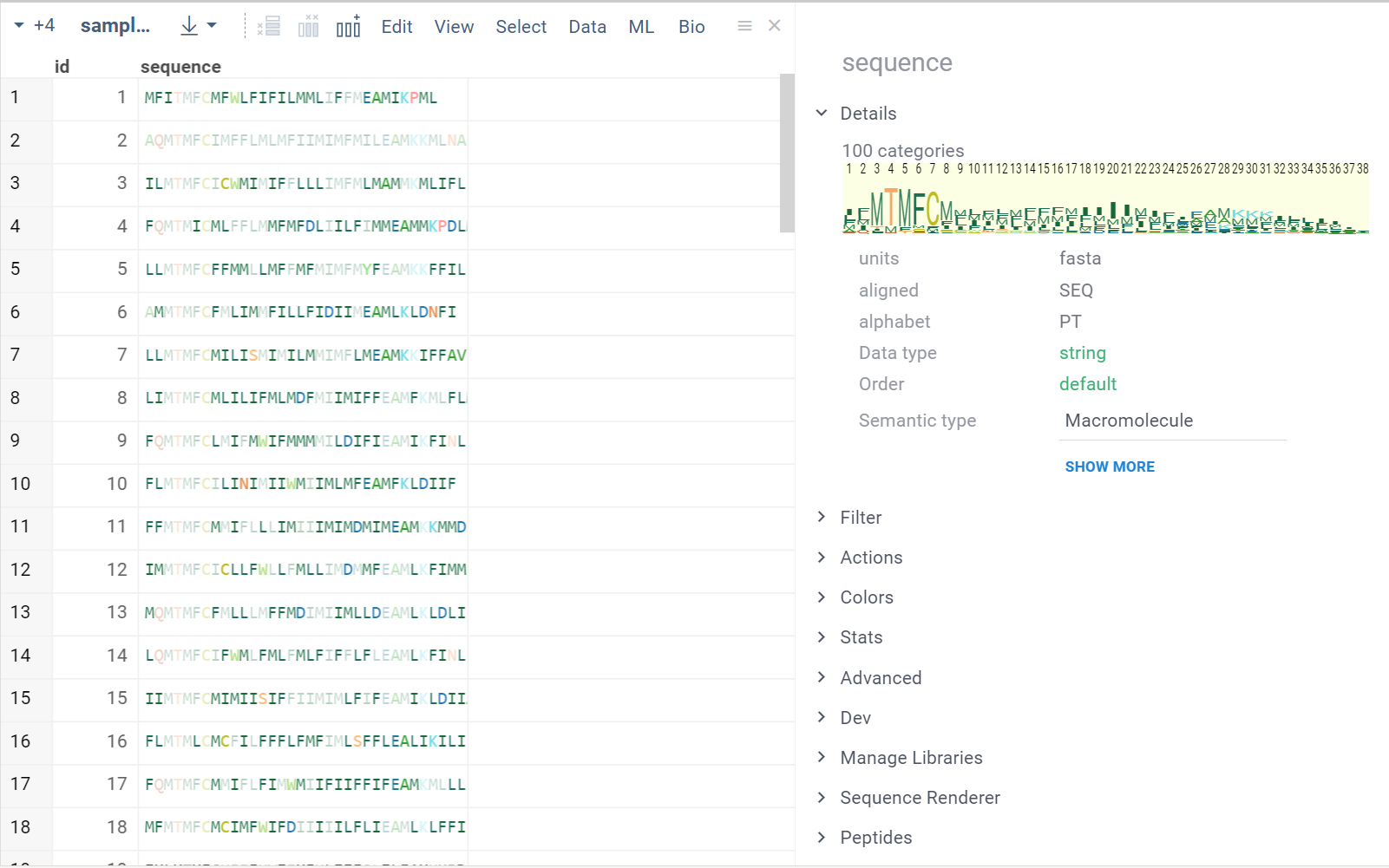
For PDB files, cells display a preview of the 3D structure. When you click a cell, a separate viewer opens up, allowing you to rotate, zoom in, or switch the color scheme.
Macromolecule aware viewers
Datagrok viewers recognize and display macromolecules. The majority of the viewers were built from scratch to take advantage of Datagrok's in-memory database, enabling seamless access to the same data across all viewers. Viewers also share a consistent design and usage patterns. Any action taken on one viewer, such as hovering, selecting, or filtering, is automatically applied to all other viewers, creating an interconnected system ideal for exploratory data analysis.
Macromolecule-specific viewers include sequence logo, 3D structure viewers (biostructure and NGL viewer), and sequence tree viewers. Examples of general-purpose viewers that can be used to analyze biological data include a scatterplot, a network diagram, a tile viewer, a bar chart, a form viewer, and trellis plot, and others.
Examples
- Spreadsheet
- Scatterplot
Composition analysis for MSA.
Activity cliffs analysis using a scatterplot.
All viewers can be saved as part of the layout or a dashboard. Some viewers offer built-in statistics.
To learn how to use viewers to explore data, complete this tutorial or visit the Visualize section of our documentation.
You can add custom viewers.
Analyzing docking results
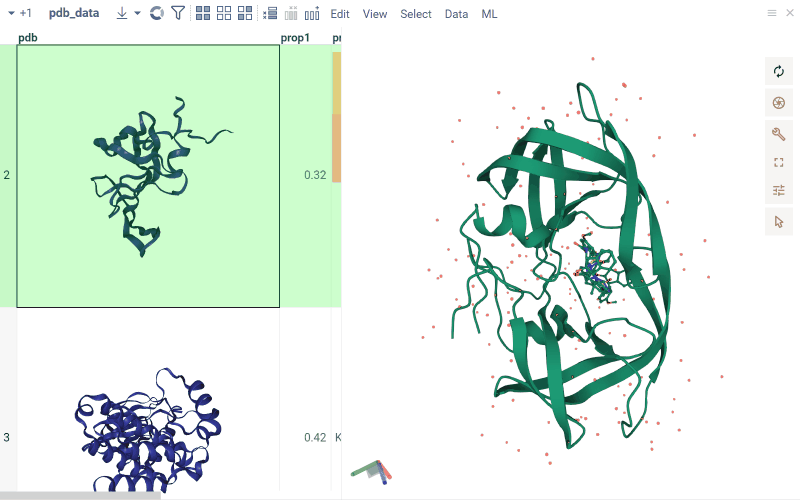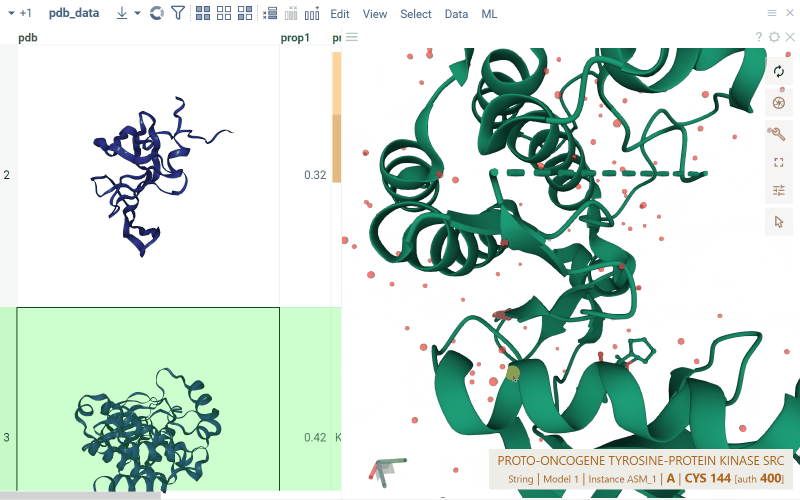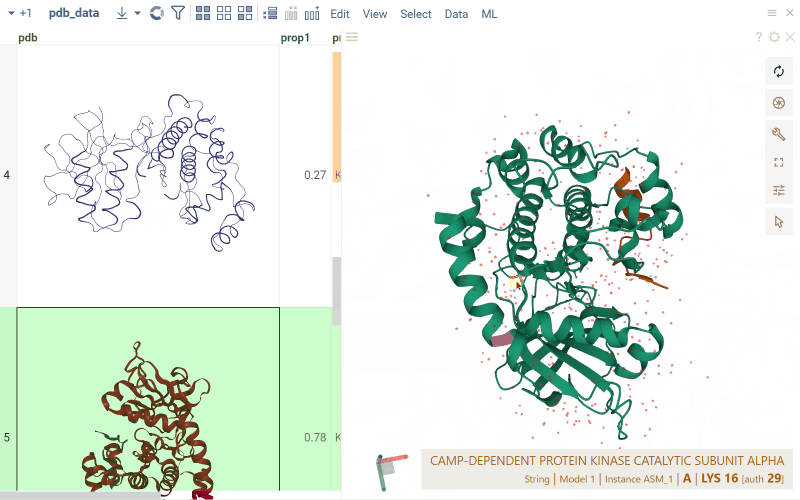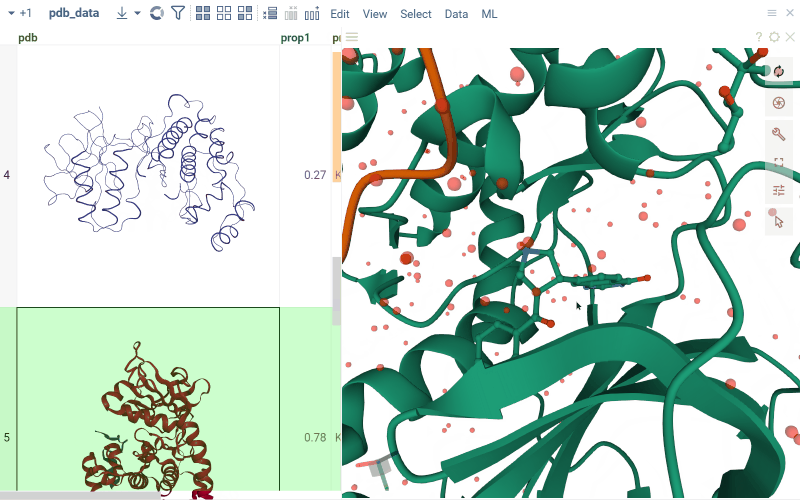
To explore the binding interactions between small ligand molecules and biological structures, you can use either the NGL or biostructure viewer. Both viewers visualize the docked ligands in their spatial context and let you examine their orientation and positioning within the binding site.
How to use
Prerequisites: Prepare the simulation data in two separate files:
- File 1: Contains the structure of a macromolecule in a supported format, such as PDB.
- File 2: Contains the simulation results of the position of small molecules relative to the structure.
To visualize docked ligands, follow these steps:
- Open the table with ligands (File 2) in Datagrok.
- Add either the NGL or biostructure viewer.
- In the viewer, drop the file with the macromolecule structure (File 1).
You can interact with the ligands as follows:
- To visualize docking for a particular ligand, click it. You can visualize multiple ligands simultaneously by selecting or hovering over rows.
- If one ligand is selected, it will be visualized with a full-color ball+stick representation.
- For multiple ligands:
- The current row ligand is shown in green.
- The mouse-over row ligand is shown in light gray.
- The selected row(s) ligands is shown in orange.
You can further analyze the ligand data by applying filters, allowing you to focus on specific subsets based on criteria such as binding energy estimates or scoring functions. You can also cluster the ligands based on one or more parameters to identify patterns and groups within the dataset.
Sketching and editing
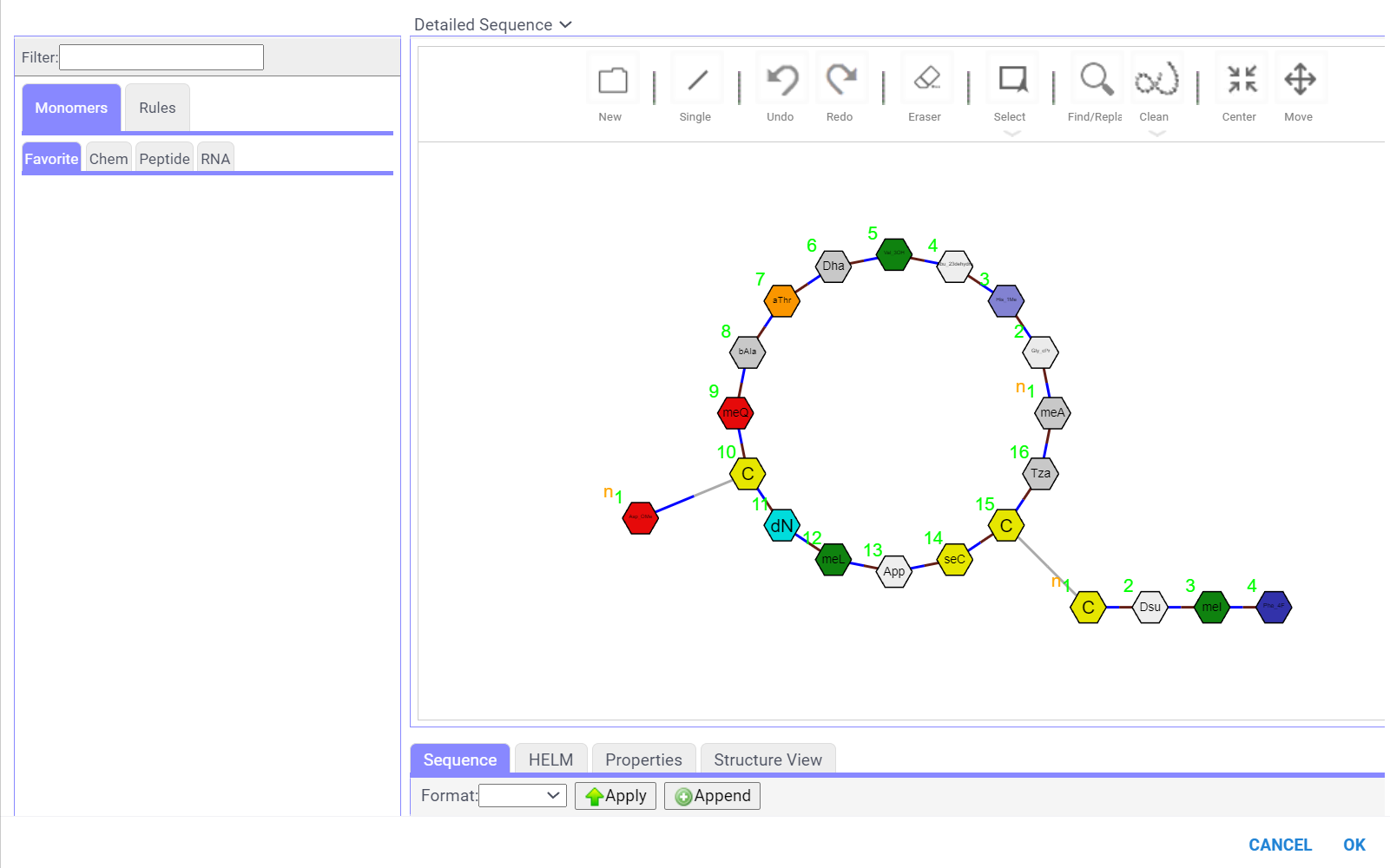
You can create and edit macromolecules:
- For DNA, RNA, and protein sequences in the linear format, you can edit the sequences.
- For HELM notation, you can add or remove monomers and modify connections. The editor supports circular and branching structures.
Searching and filtering
Datagrok offers an intuitive searching and filtering functionality for exploring datasets.
For sequence-based filtering of macromolecules, Datagrok uses integrated HELM editor for HELM notations and text-based filter for linear notations. All filters are interactive. Hovering over categories or distributions in the Filter Panel instantly highlights relevant data points across all viewers.
The search feature provides another way to analyze your dataset. The search results are presented as a new column in the table, where checkboxes indicate whether each sequence is a match or not. You can color-code the search results for quick visual profiling. In addition, like any other column in the table, this column can be used as a filter. This means you can create and apply additional filters based on the search results, facilitating further analysis and exploration of the dataset.
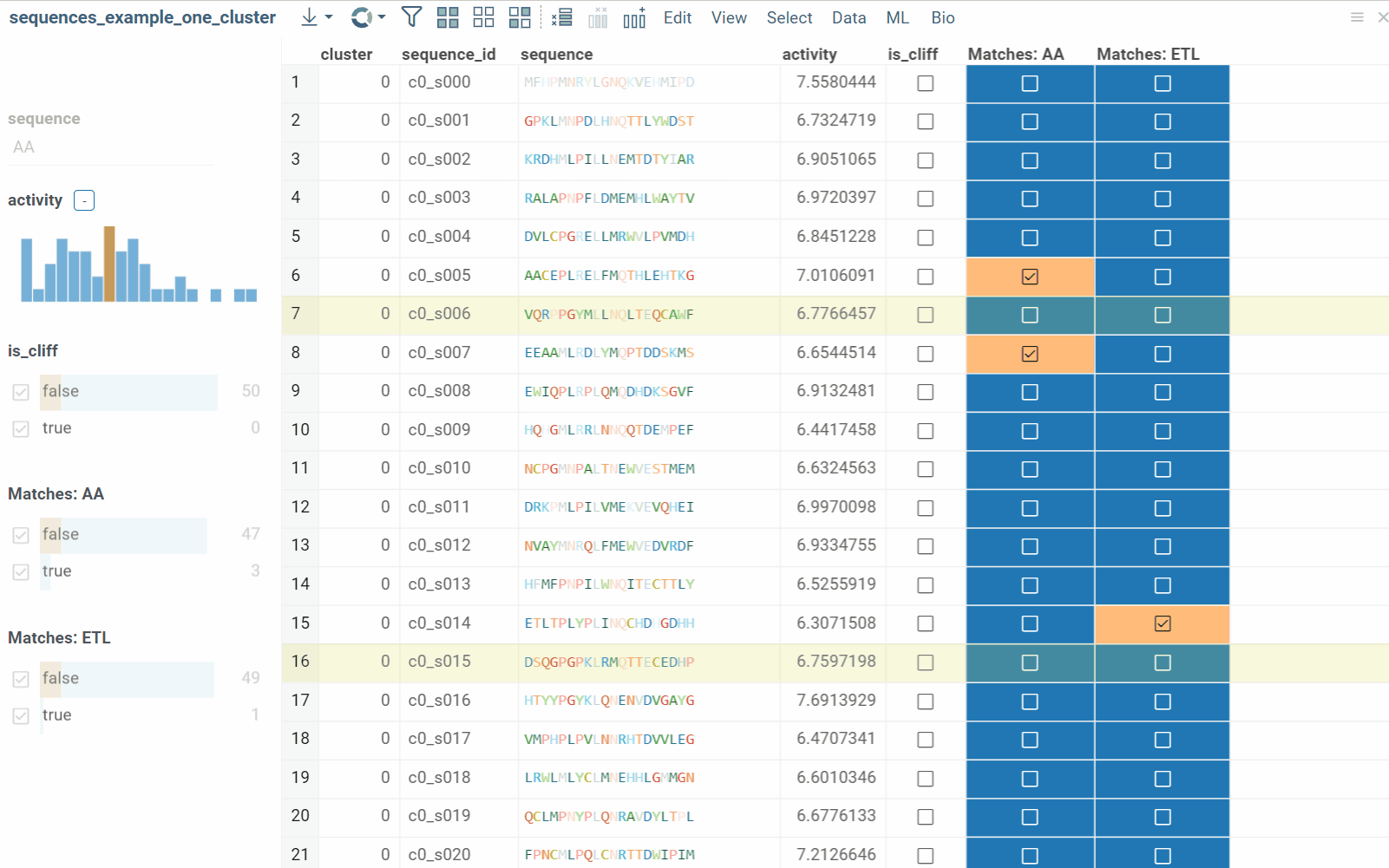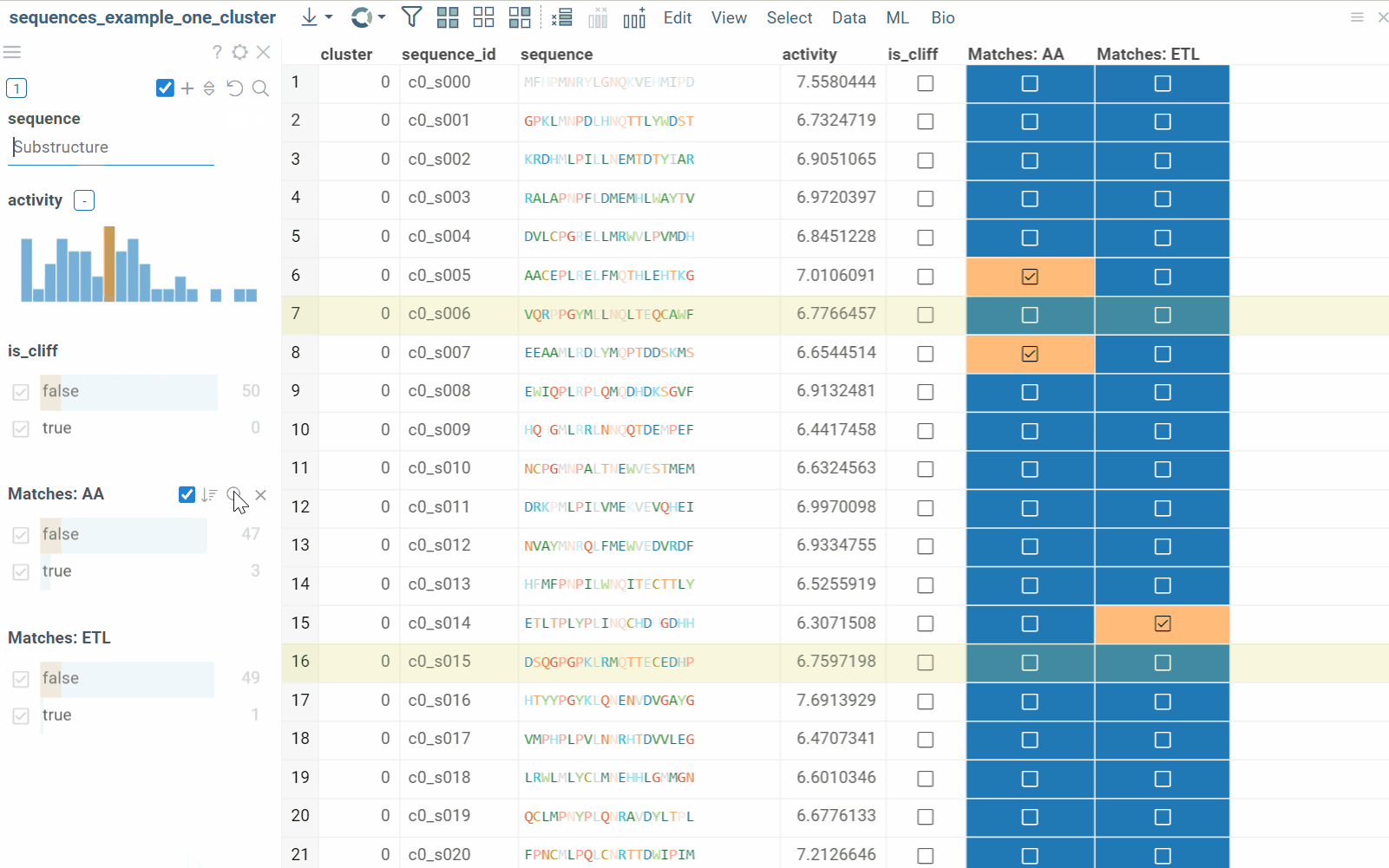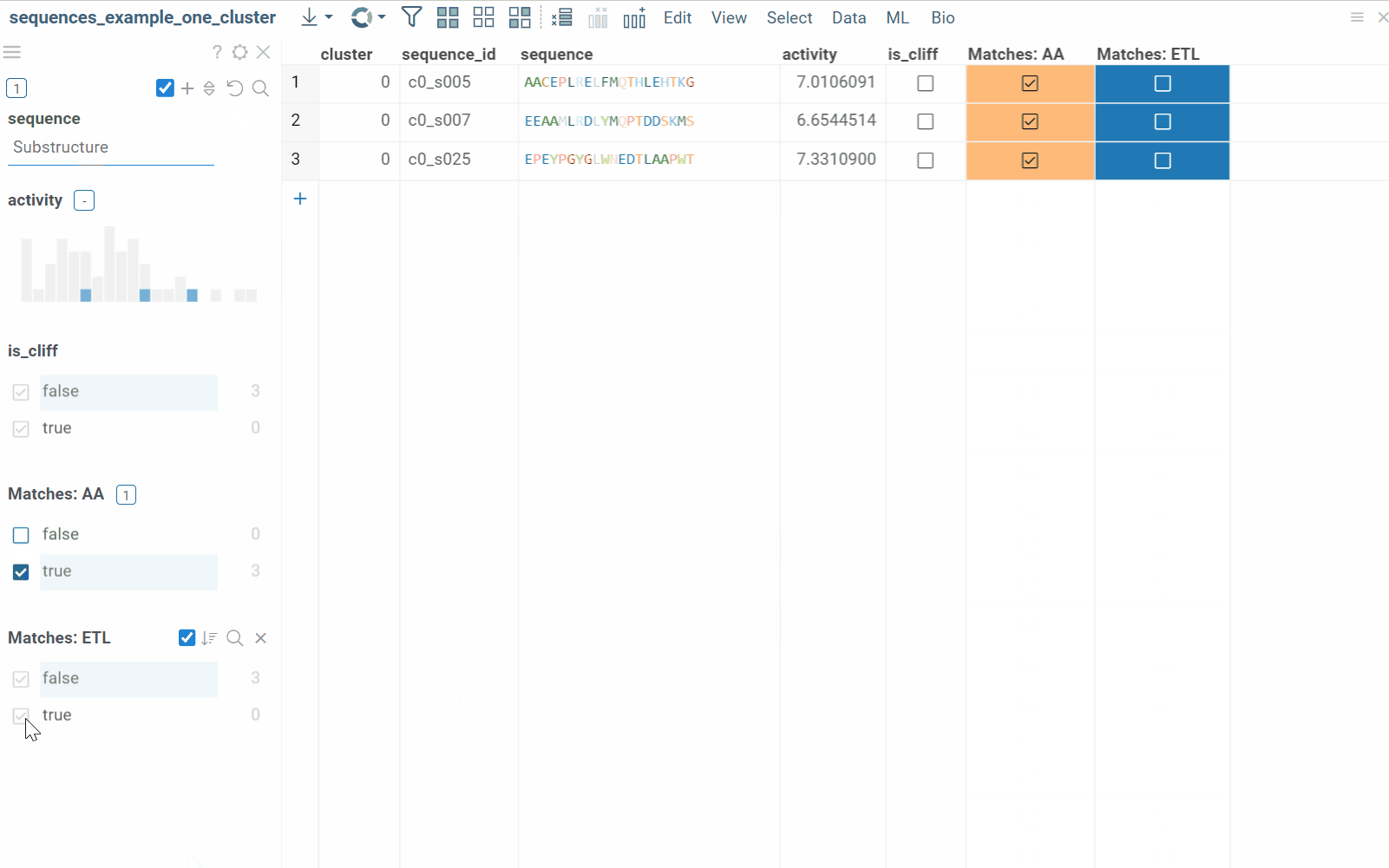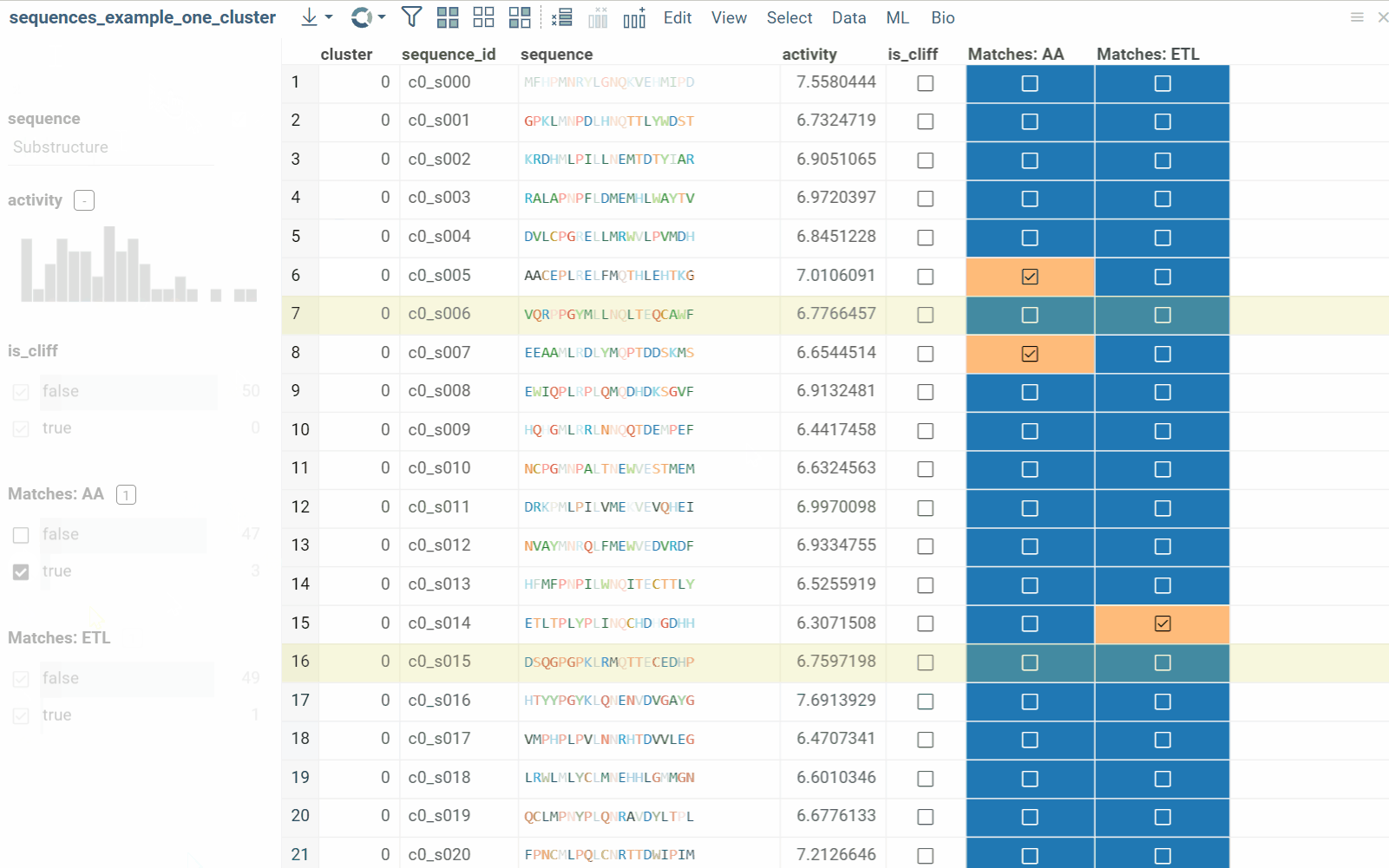
How to filter
To filter by sequence, do the following:
- On the Top Menu, click the Filter icon to open the Filter Panel. The panel shows filters for all dataset columns. By default, the subsequence filter is displayed on top but you can rearrange, add, or remove filter columns by using available controls.
- To enter a subsequence, Click to edit button.
- Once finished, click OK to apply the filter.
To clear the filter, use the checkbox provided. To remove the filter altogether, use the Close (x) icon.
How to search
To search a dataset for matching sequences, do the following:
- In the Top Menu, select Bio > Substructure Search... A dialog opens.
- In the dialog, paste or type the sequence in the field provided and click OK. A new column is added to the table.
To learn more about filtering, watch this video or read this article.
Manage monomer libraries
The default HELM monomer library is pre-installed with the Bio package. You can add your own monomer libraries using the dialog accessible from Top Menu > Bio > Manage > Monomer Libraries:
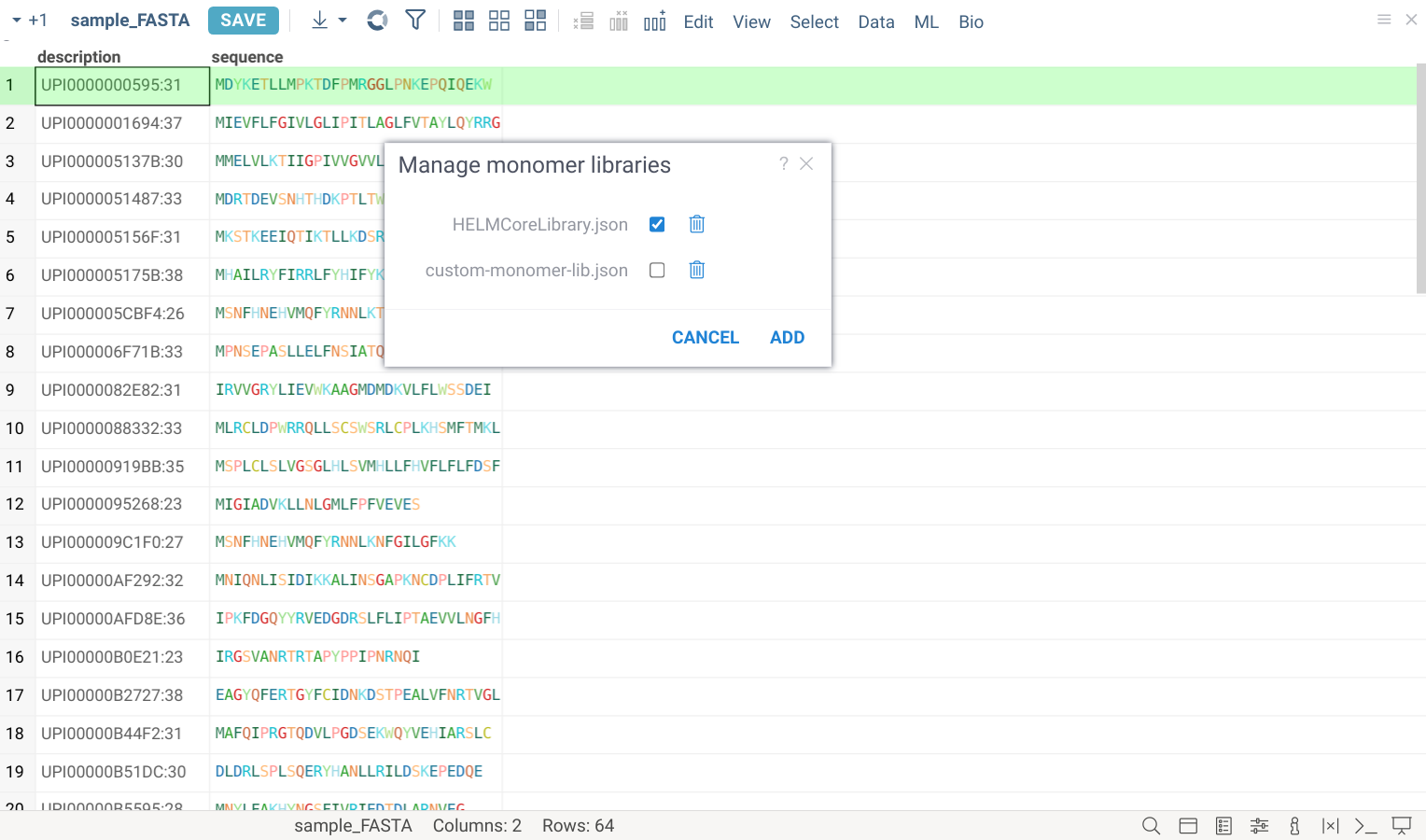
To include monomers from a library, click on a checkbox next to its name.
To add a new monomer library file, click ADD button. All monomer library files are validated against the standard HELM JSON schema and must fully conform to it. The added files will be stored under AppData/Bio/monomer-libraries in file shares.
Sequence analysis
Sequence composition
You can create a sequence logo to show the letter composition for each position in a sequence. A sequence logo is usually created from a set of aligned sequences and helps identify patterns and variations within those sequences. A common use is to visualize protein-binding sites in DNA or functional motives in proteins.
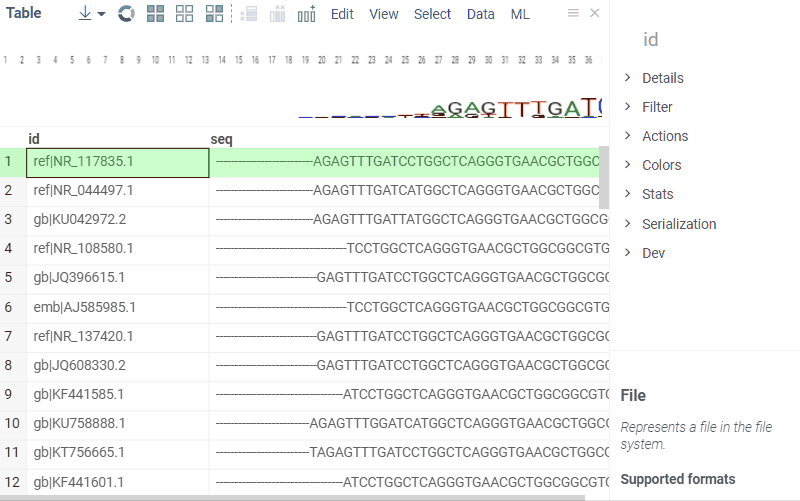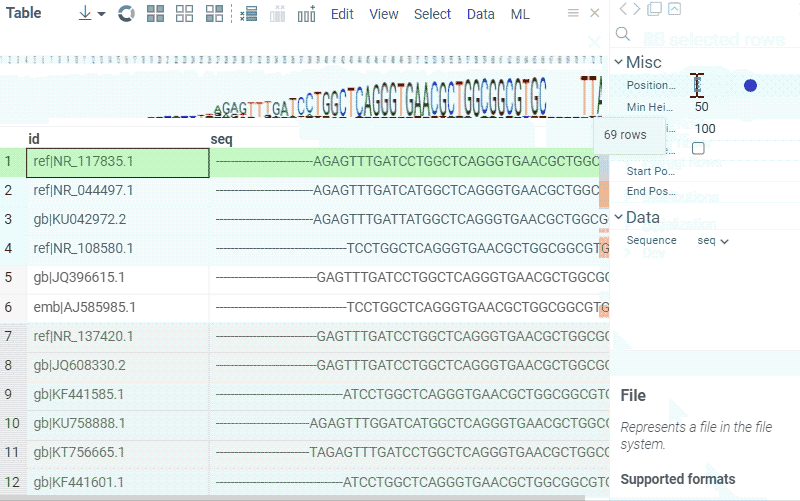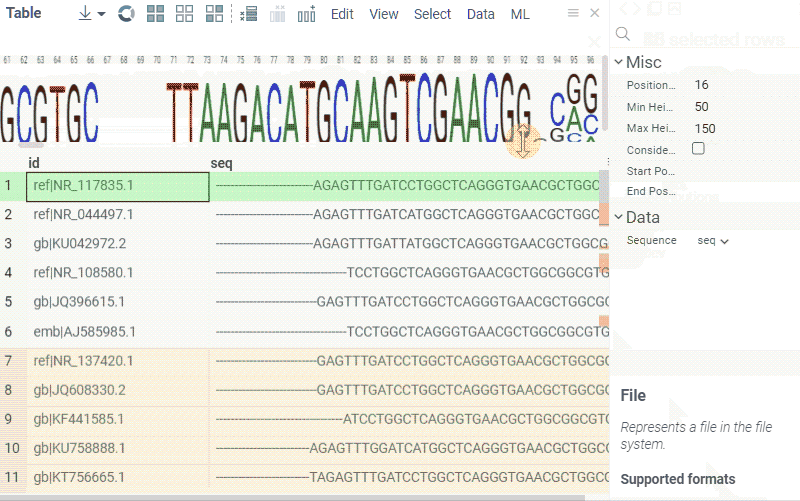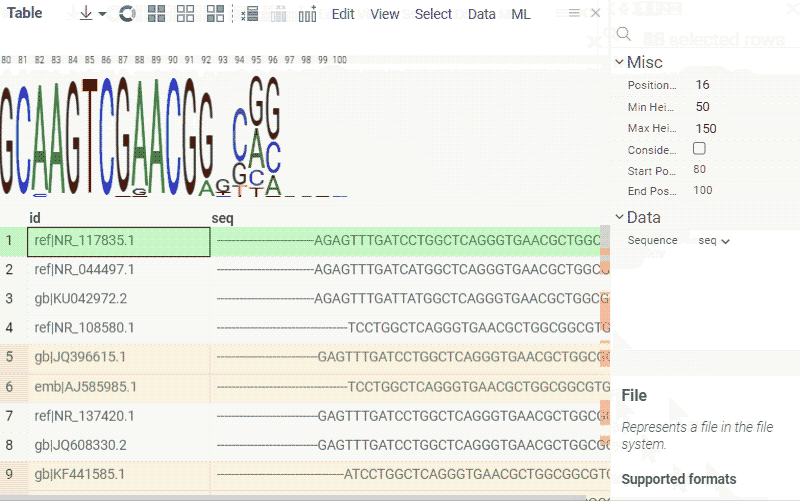
How to use
- In the Top Menu, select Bio > Composition Analysis. The sequence logo viewer is added to the Table View.
- To edit parameters, hover over the viewer's top and click the Gear icon.
Sequence space
Sequence space visualizes a collection of sequences in 2D such that similar sequences are placed close to each other (geekspeak: dimensionality reduction, tSNE, UMAP, distance functions). This allows to identify clusters of similar sequences, outliers, or patterns that might be difficult to detect otherwise. Results are visualized on the interactive scatterplot.
Sequence space analysis is particularly useful for separating groups of sequences with common motifs, such as different variants of complementarity-determining regions (CDRs) for antibodies.
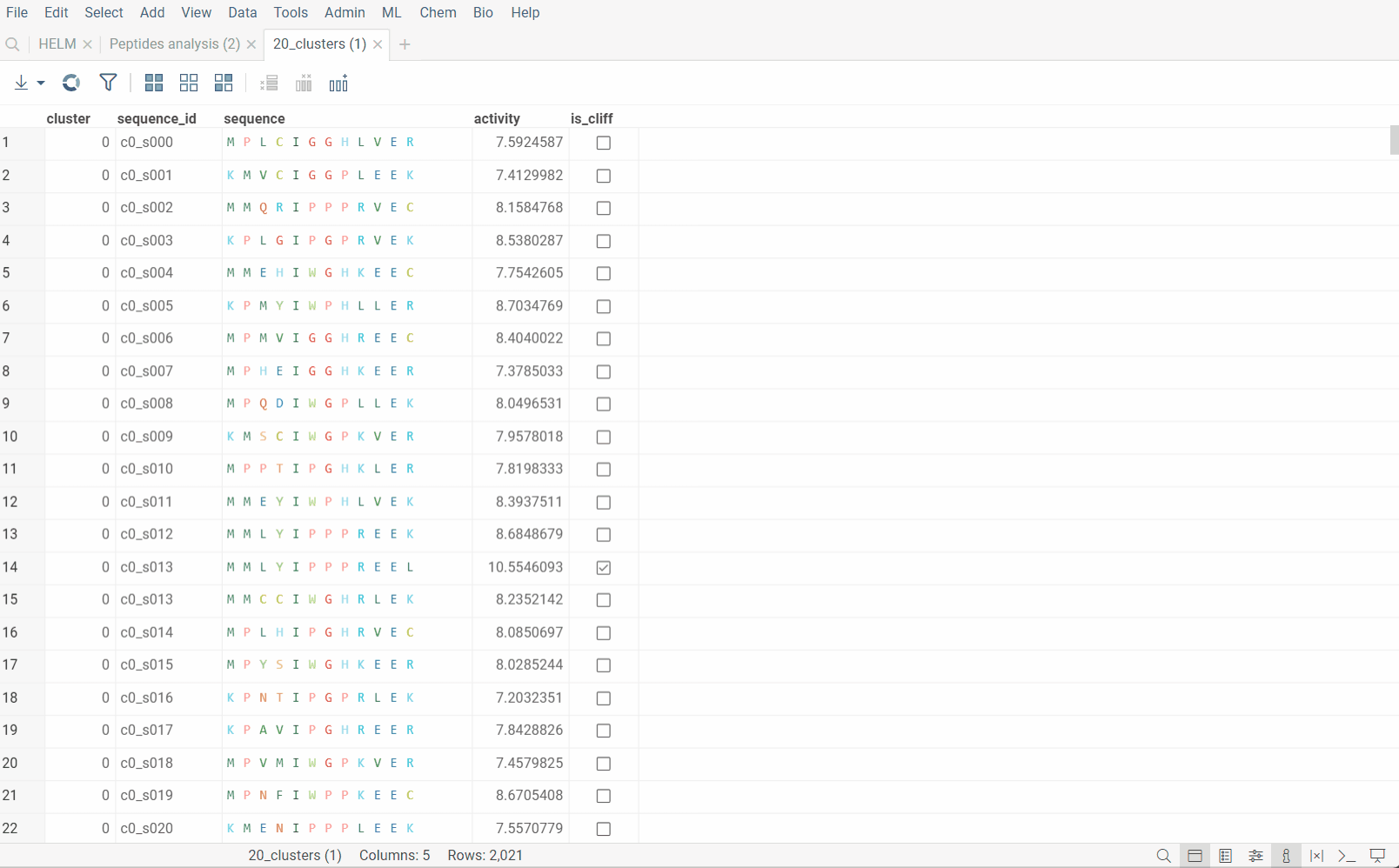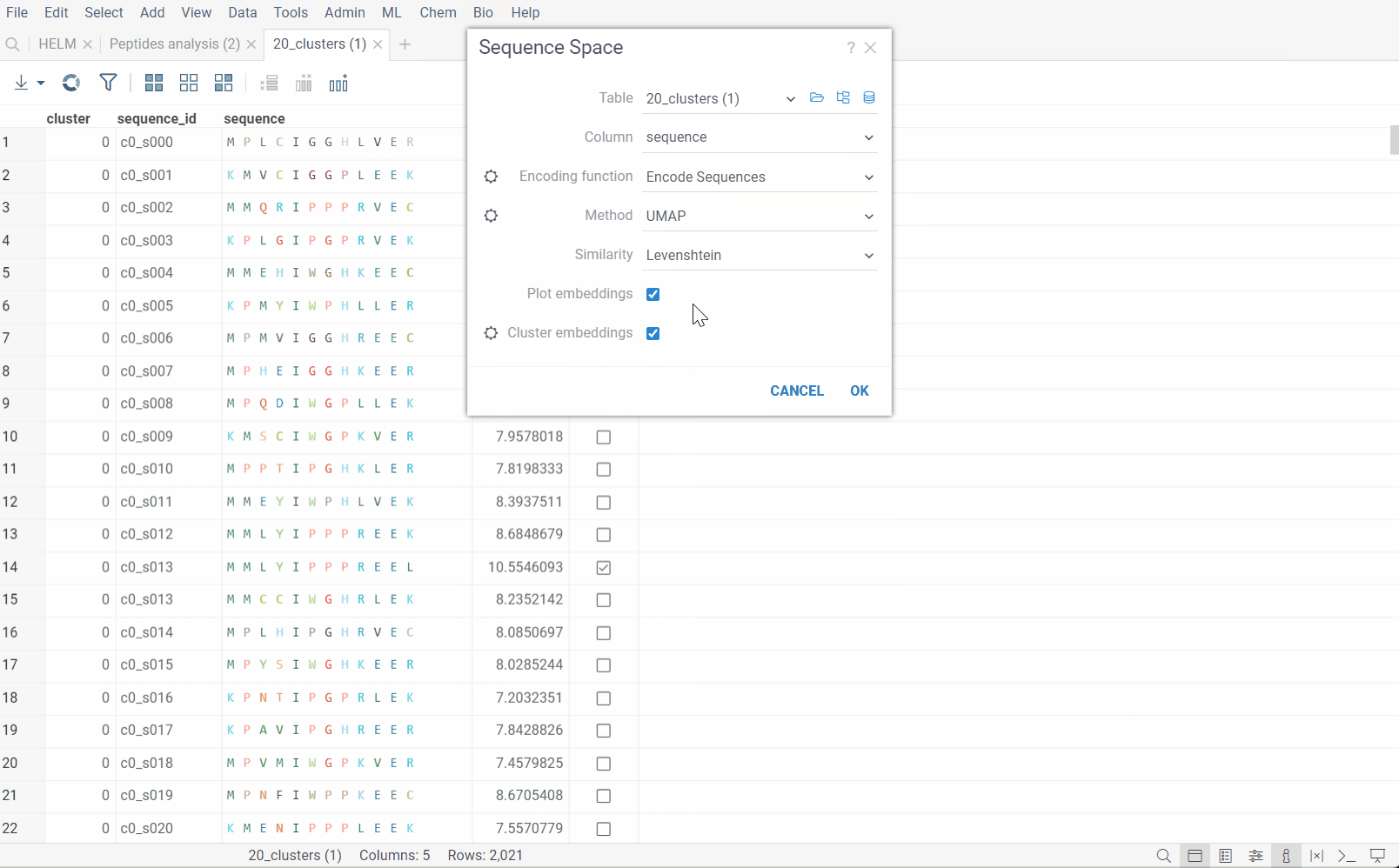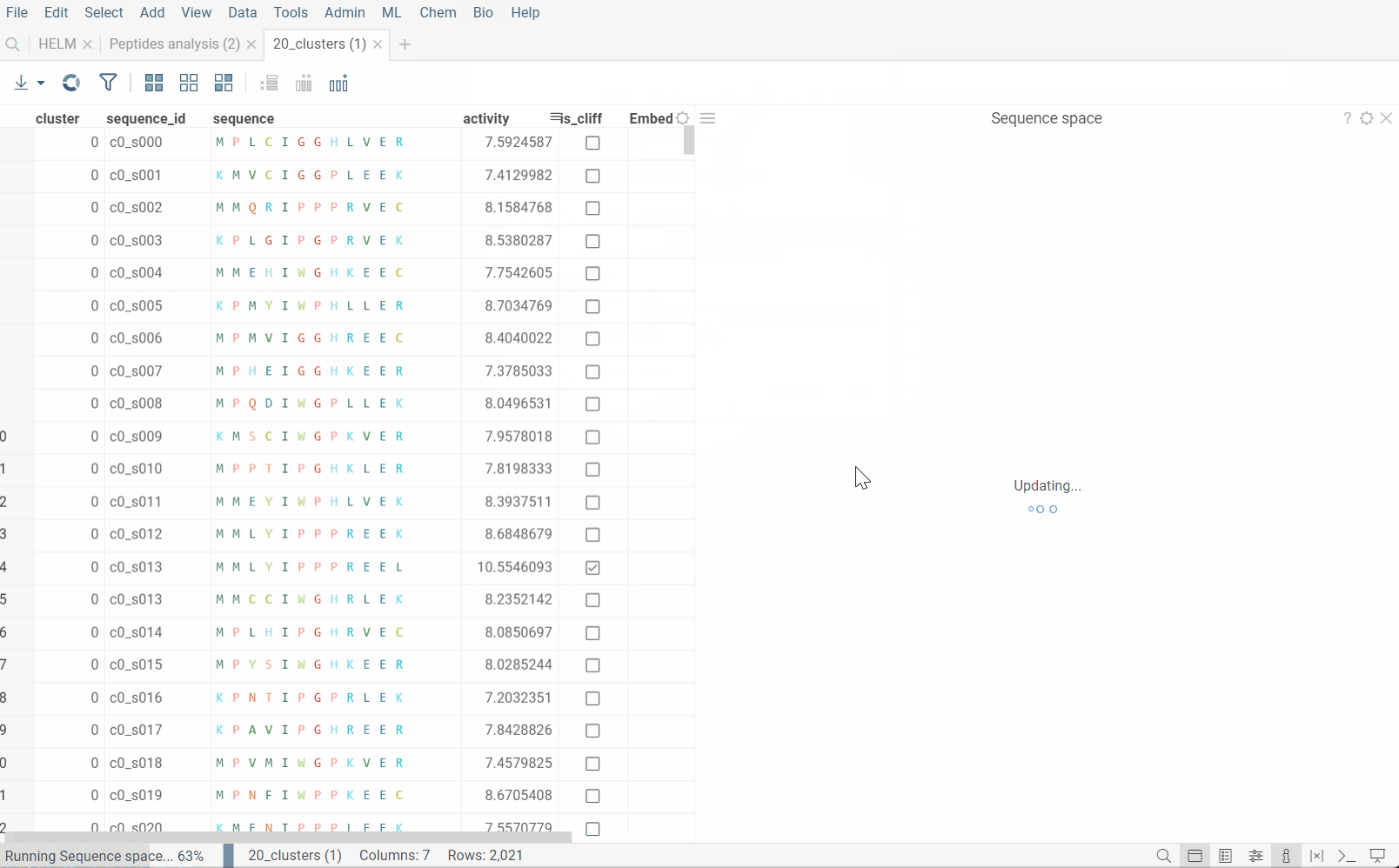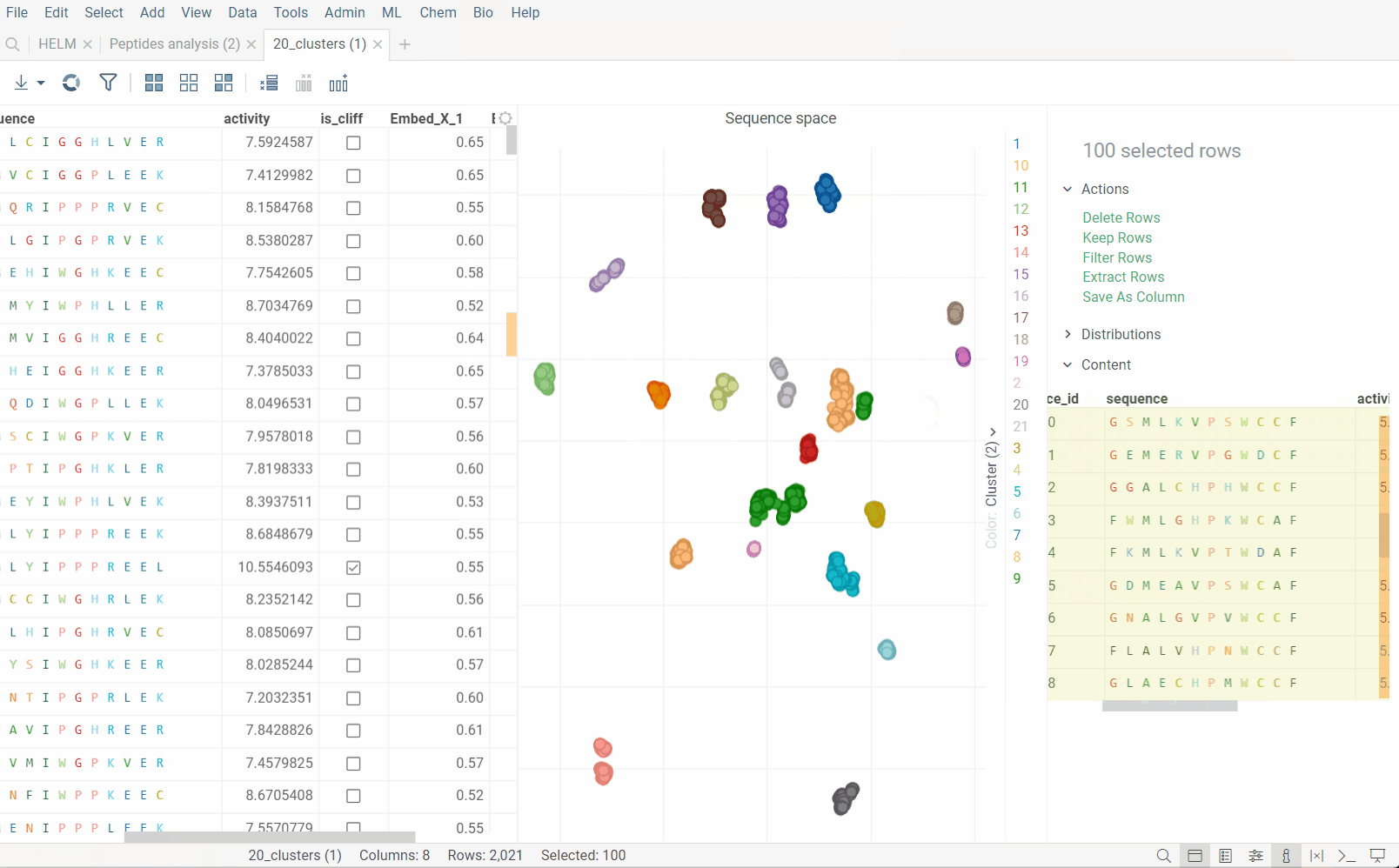
How to use
Go to the Top Menu and select Bio > Analyse > Sequence Space... This opens a Sequence Space parameter dialog.
The dialog has the following inputs:
-
Table: The table containing the column of sequences.
-
Column: The column containing the sequences.
-
Encoding function: The encoding function that will be used for pre-processing of sequences. For non-helm notation sequences, only one encoding function is available, that will encode them in single charachter form and calculate the substitution matrix for each individual monomer. For Helm sequences, apart from prior function, another one is offered that will use chemical fingerprint distances between each macromolecule to calculate distance matrix. The
Encode sequencesfunction has 3 parameter which you can adjust using the gear (⚙️) button next to the encoding function selection:- Gap open penalty: The penalty for opening a gap in the alignment (used for Needleman-Wunsch algorythm).
- Gap extend penalty: The penalty for extending a gap in the alignment (used for Needleman-Wunsch algorythm).
- Fingerprint type: The type of molecular fingerprints that will be used to generate monomer substitution matrix.
-
Method: The dimensionality reduction method that will be used. The options are:
- UMAP: UMAP is a dimensionality reduction technique that can be used for visualisation similarly to t-SNE, but also for general non-linear dimension reduction.
- t-SNE: t-SNE is a machine learning algorithm for dimensionality reduction developed by Geoffrey Hinton and Laurens van der Maaten. It is a nonlinear dimensionality reduction technique that is particularly well-suited for embedding high-dimensional data into a space of two or three dimensions, which can then be visualized in a scatter plot.
Other parameters for dimensionality reduction method can be accessed through the gear (⚙️) button next to the method selection.
-
Similarity: The similarity/distance function that will be used to calculate pairwise distances. The options are:
- Needleman-Wunsch: Needleman-Wunsch is a dynamic programming algorithm that performs a global alignment on two sequences. It is commonly used in bioinformatics to align protein or nucleotide sequences.
- Hamming: Hamming distance is a metric for comparing two macromolecules of same length. Hamming distance is the number of positions in which the two monomers are different.
- Monomer chemical distance: Similar to Hamming distance, but instead of penalizing the missmatch of monomers with -1, the penalty will be based on the chemical distance between the two monomers. The chemical distance is calculated using the chemical fingerprint of the monomers.
- Levenshtein: Levenshtein distance is a string metric for measuring the difference between two sequences. Informally, the Levenshtein distance between two sequences is the minimum number of single-monomer edits (insertions, deletions or substitutions) required to change one sequence into the other.
-
Plot embeddings: If checked, the plot of the embeddings will be shown after the calculation is finished.
-
Postprocessing: The postprocessing function that will be applied to the resulting embeddings. The options are:
- None: No postprocessing will be applied.
- DBSCAN: The DBSCAN algorithm groups together points that are closely packed together (points with many nearby neighbors), marking as outliers points that lie alone in low-density regions (whose nearest neighbors are too far away). The DBSCAN algorithm has two parameters that you can adjust through the gear (⚙️) button next to the postprocessing selection:
- Epsilon: The maximum distance between two points for them to be considered as in the same neighborhood.
- Minimum points: The number of samples (or total weight) in a neighborhood for a point to be considered as a core point. This includes the point itself.
- Radial Coloring: The radial coloring function will color the points based on their distance from the center of the plot. The color will be calculated as a gradient from the center to the border of the plot.
WebGPU (experimental)
WebGPU is an experimental feature that allows you to use the GPU for calculations in browser. We have implemented the KNN graph generation (with support to all simple and non-trivial distance functions like Needleman-Wunsch, Levenstain, etc.) and UMAP algorithms in webGPU, which can be enabled in the dimensionality reduction dialog. This can speed up the calculations significantly, especially for large datasets, up to 100x. This option can be found in the gear (⚙️) button next to the method selection (UMAP).
Please note, that webGPU is still considered as experimental feature, and for now only works in Chrome or Edge browsers (although it is planned to be supported in Firefox and Safari in the future). If webGPU is not supported in your browser, this checkbox will not appear in the dialog. To make sure that your opperating system gives browser access to correct(faster) GPU, you can check the following:
- Go to settings and find display settings
- Go to Graphics settings.
- In the list of apps, make sure that your browser is set to use high performance GPU.
Hierarchical clustering
Hierarchical clustering groups sequences into an interactive dendrogram. In a dendrogram, distance to the nearest common node represents the degree of similarity between each pair of sequences. Clusters and their sizes can be obtained by traversing the trunk or branches of the tree and deciding at which level to cut or separate the branches. This process lets you identify different clusters based on the desired level of similarity or dissimilarity between data points.
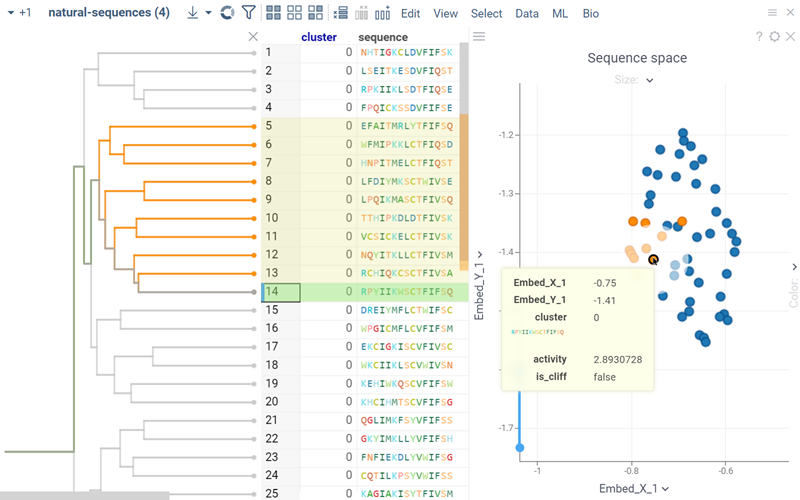
How to use
To add a dendrogram viewer, do the following:
- In the Top Menu, select Bio > Hierarchical clustering. A dialog opens.
- In the dialog, select the parameters and click OK to add the dendrogram to the Table View.
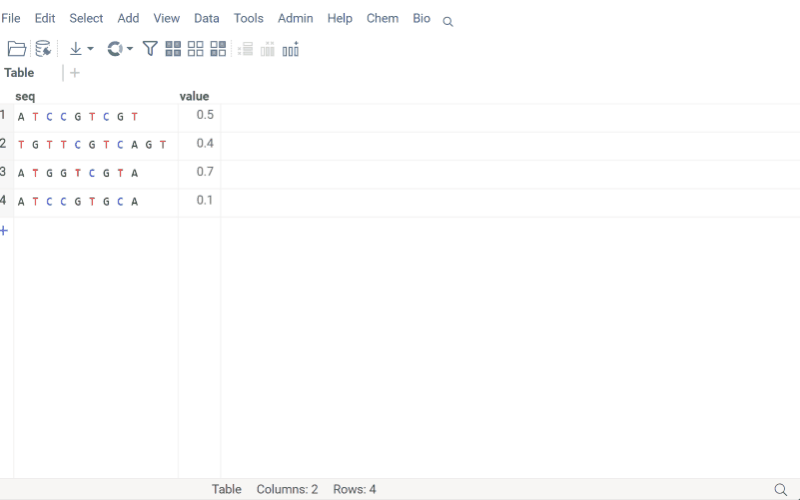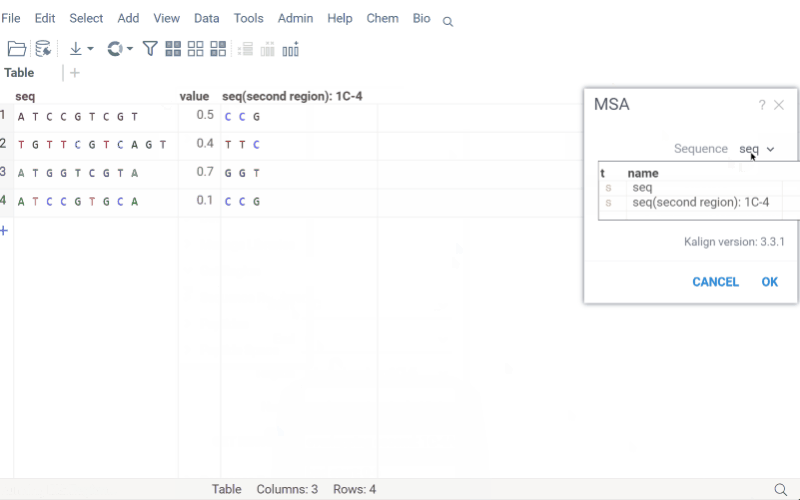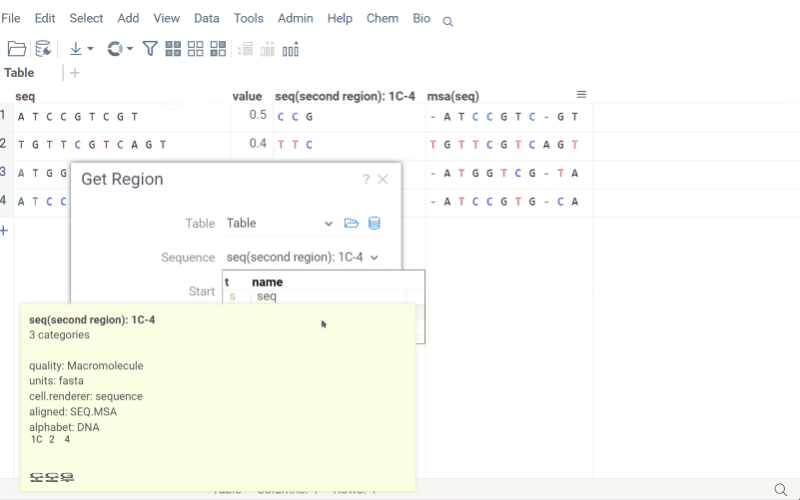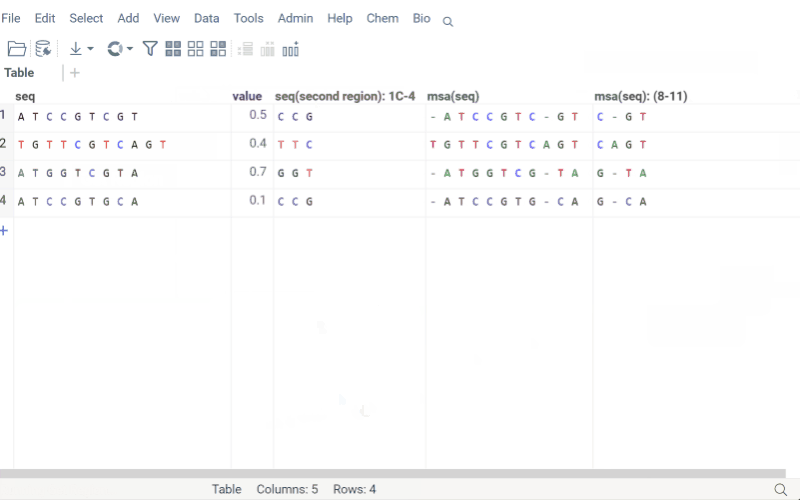
Multiple sequence alignment (MSA)
Multiple Sequence Alignment aligns sequences for macromolecules in both FASTA and HELM formats. For DNA, RNA, and natural peptides, we use KAlign, which can be modified to work with custom substitution matrices for sequences. For non-natural sequences, we use PepSeA, which enables alignment of multiple linear peptide sequences in HELM notation, with lengths of up to 256 non-natural amino acids.
How to use
To perform MSA, do the following:
- In the Top Menu, select Bio > MSA... . A dialog opens.
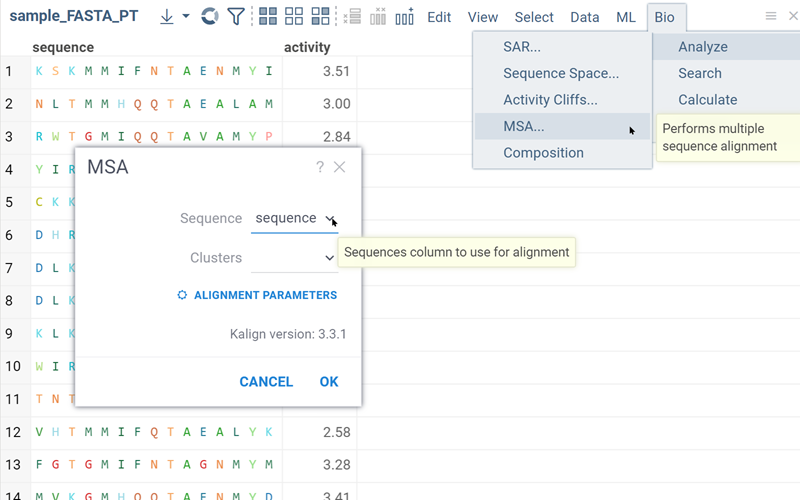
- In the dialog, select the sequence column (Sequence) and set other parameters. If your data has been clustered already, you can align sequences only within the same cluster. To do so, specify a column containing clusters (Cluster).
- Click OK to execute. A new column containing the aligned sequences is added to the table.
Sequence-Activity relationship analysis
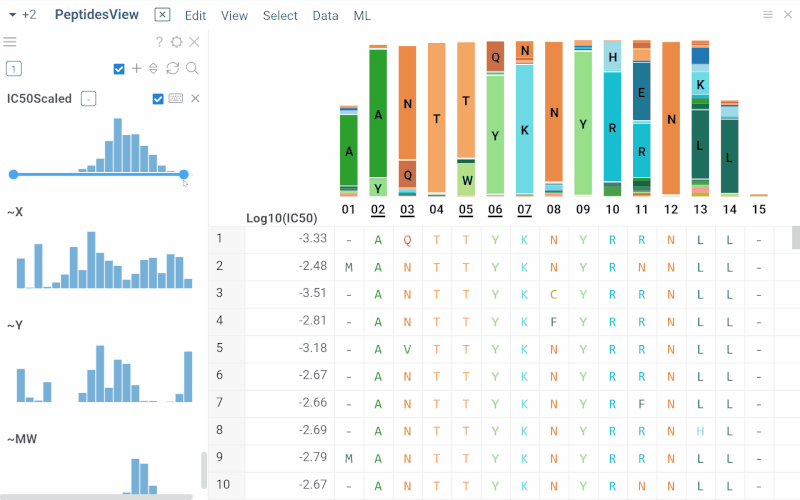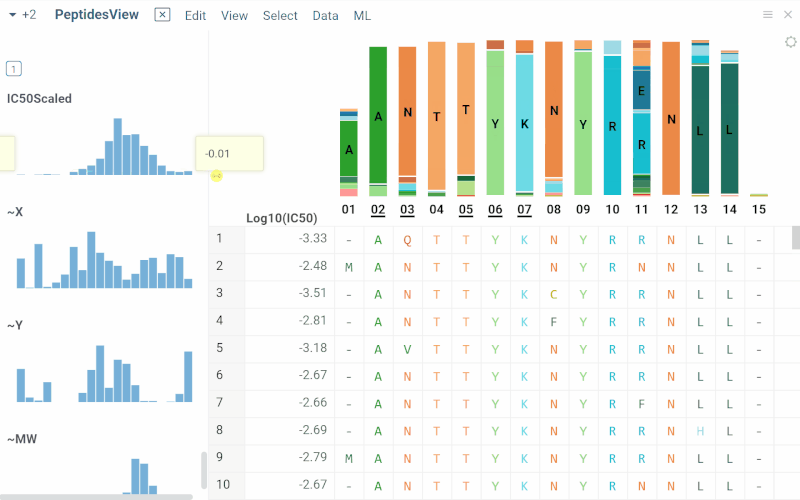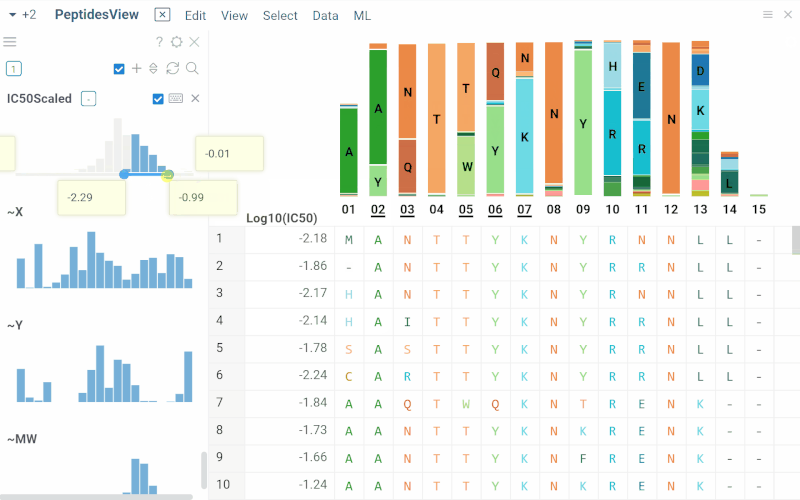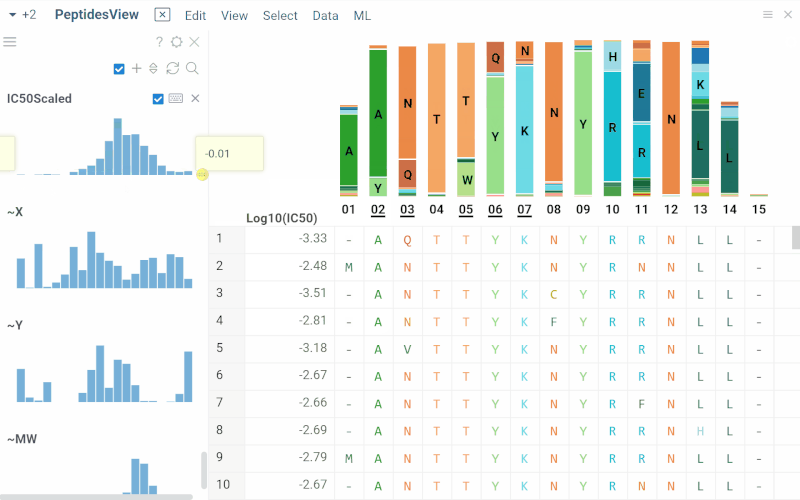
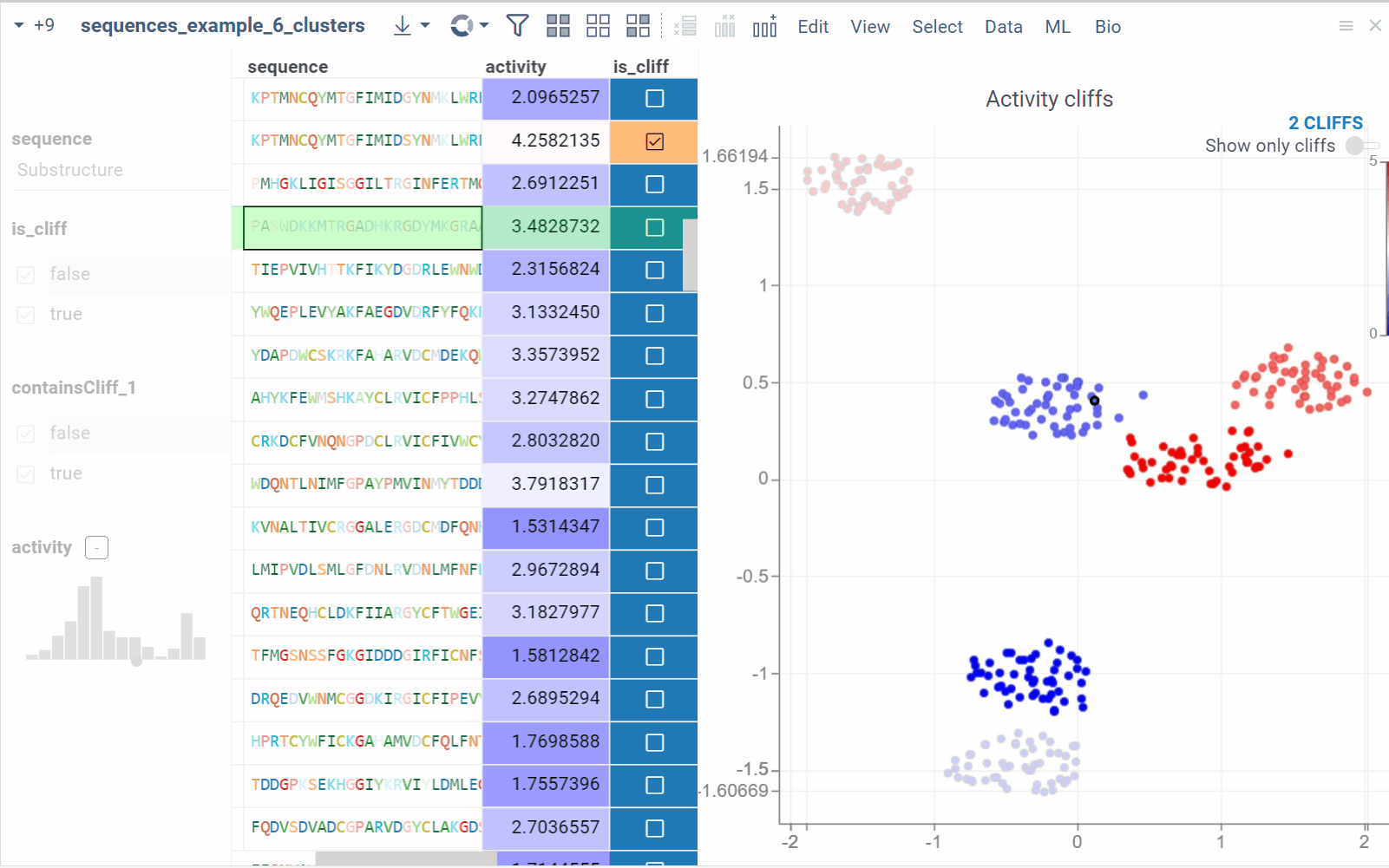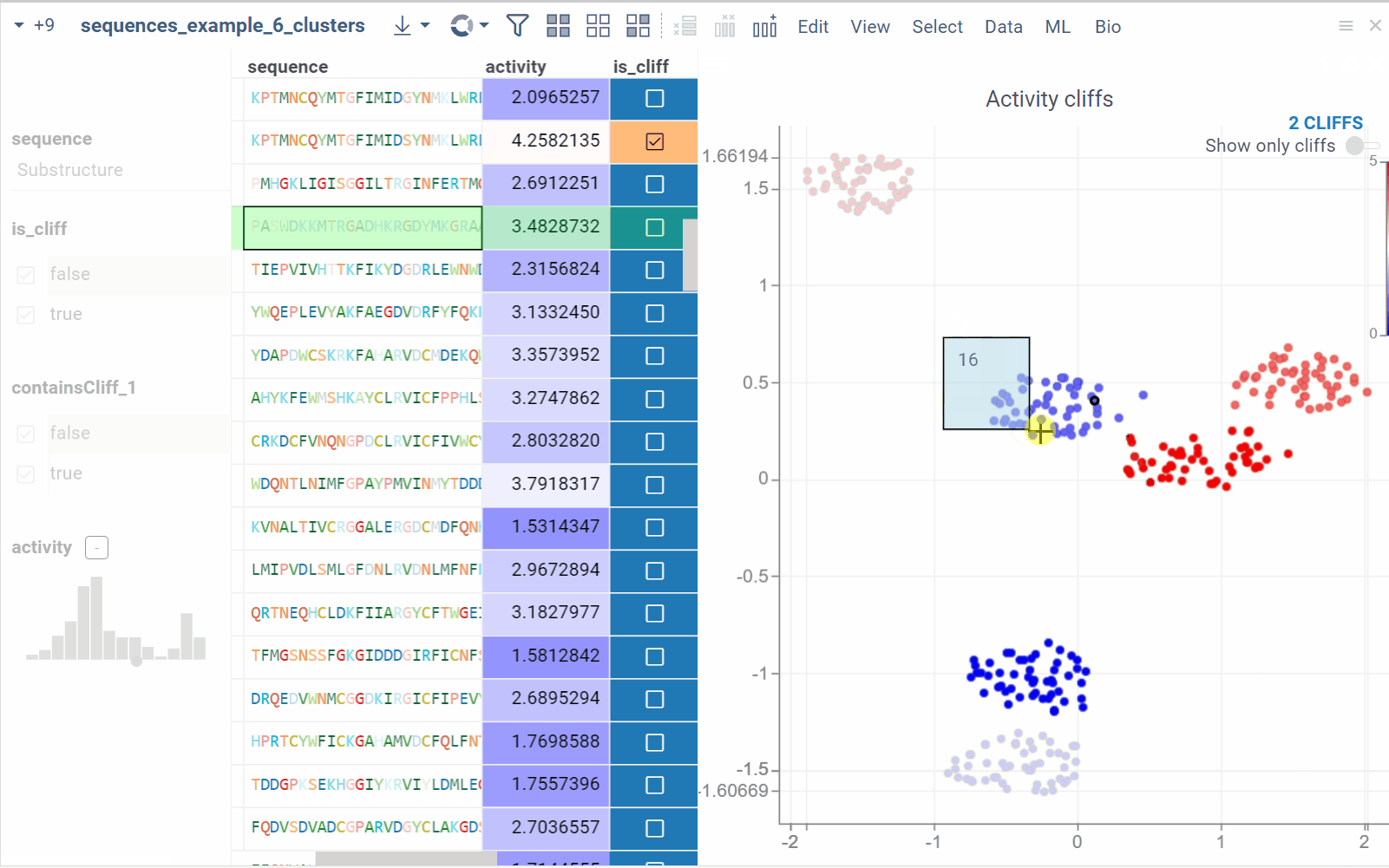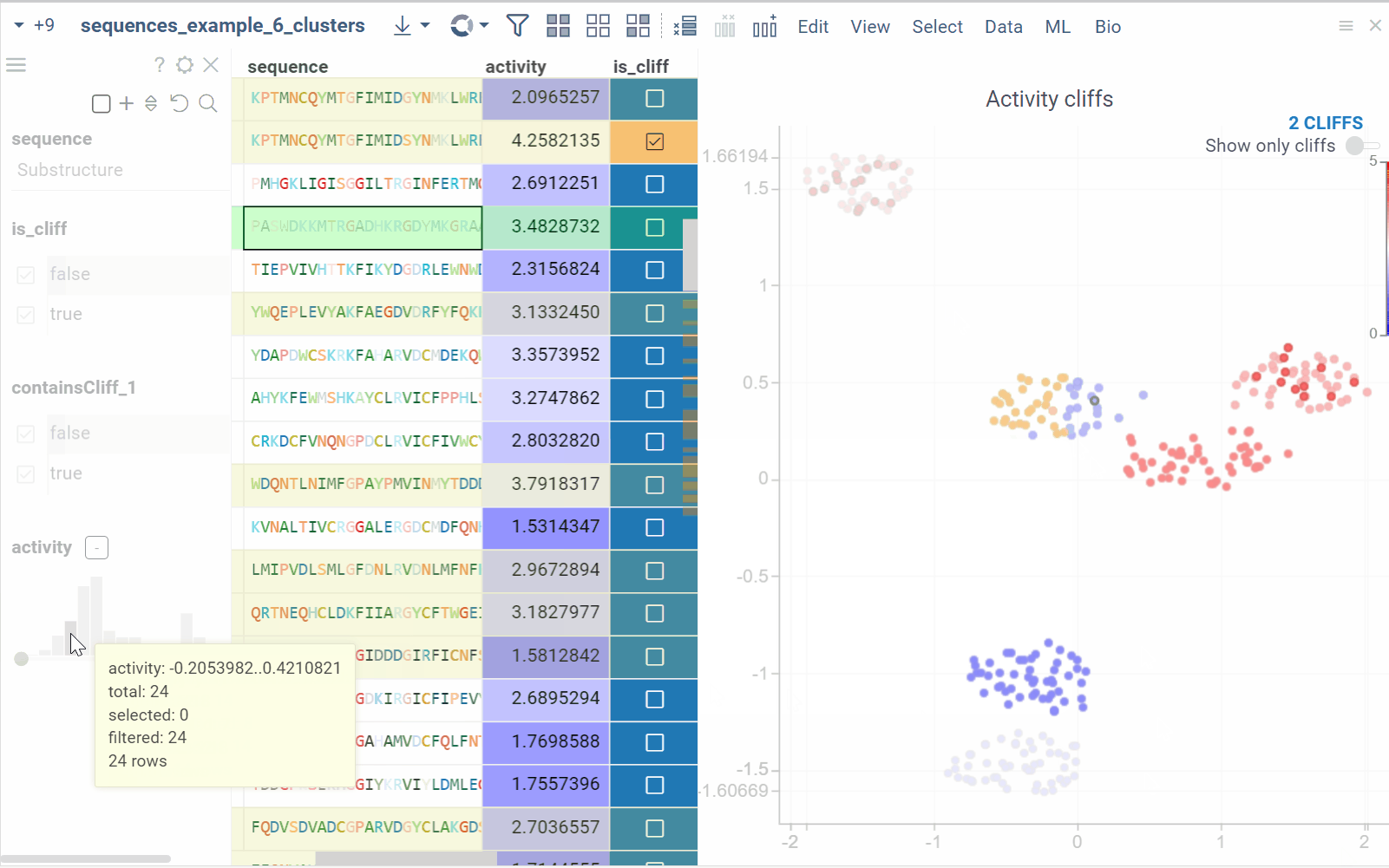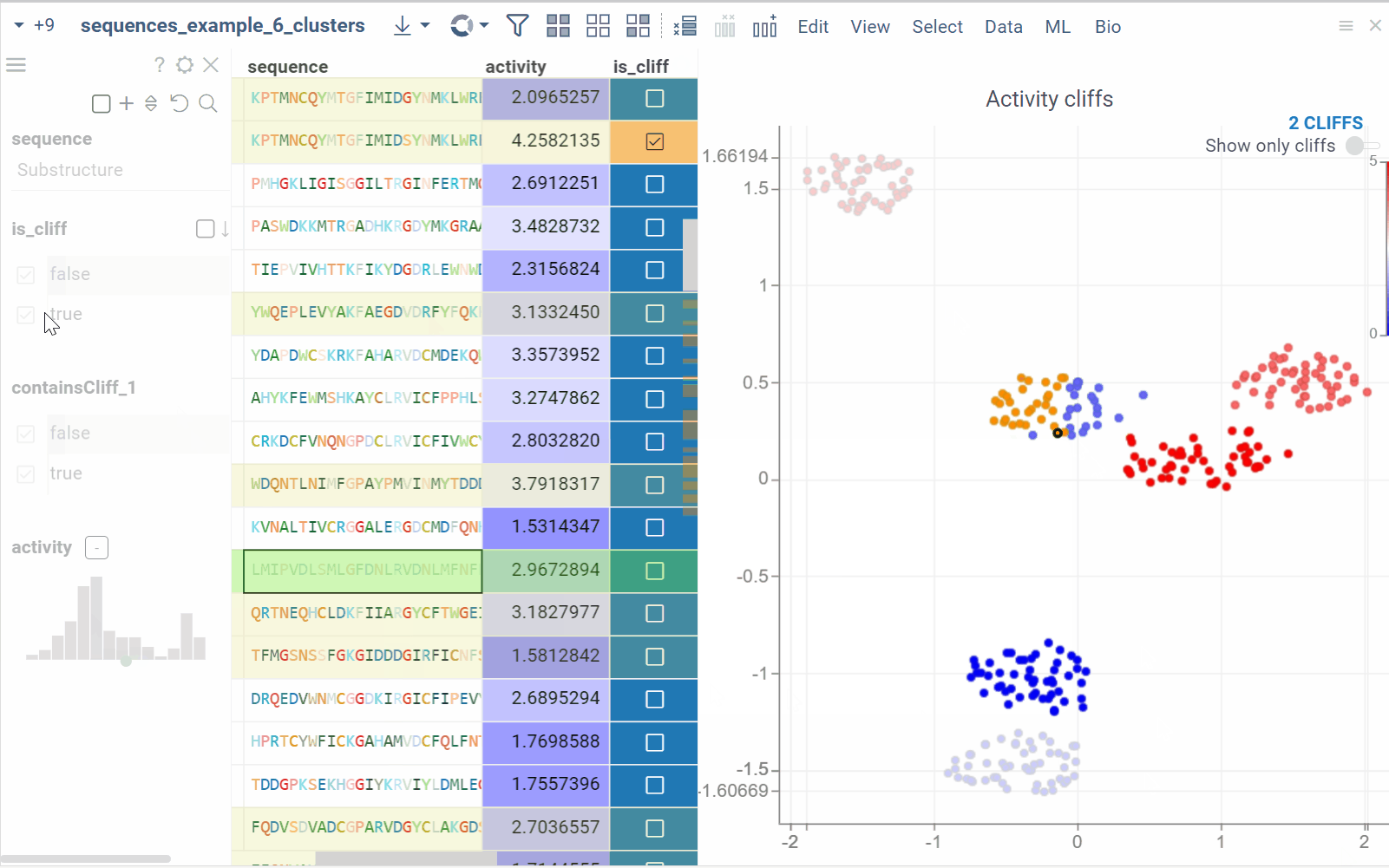
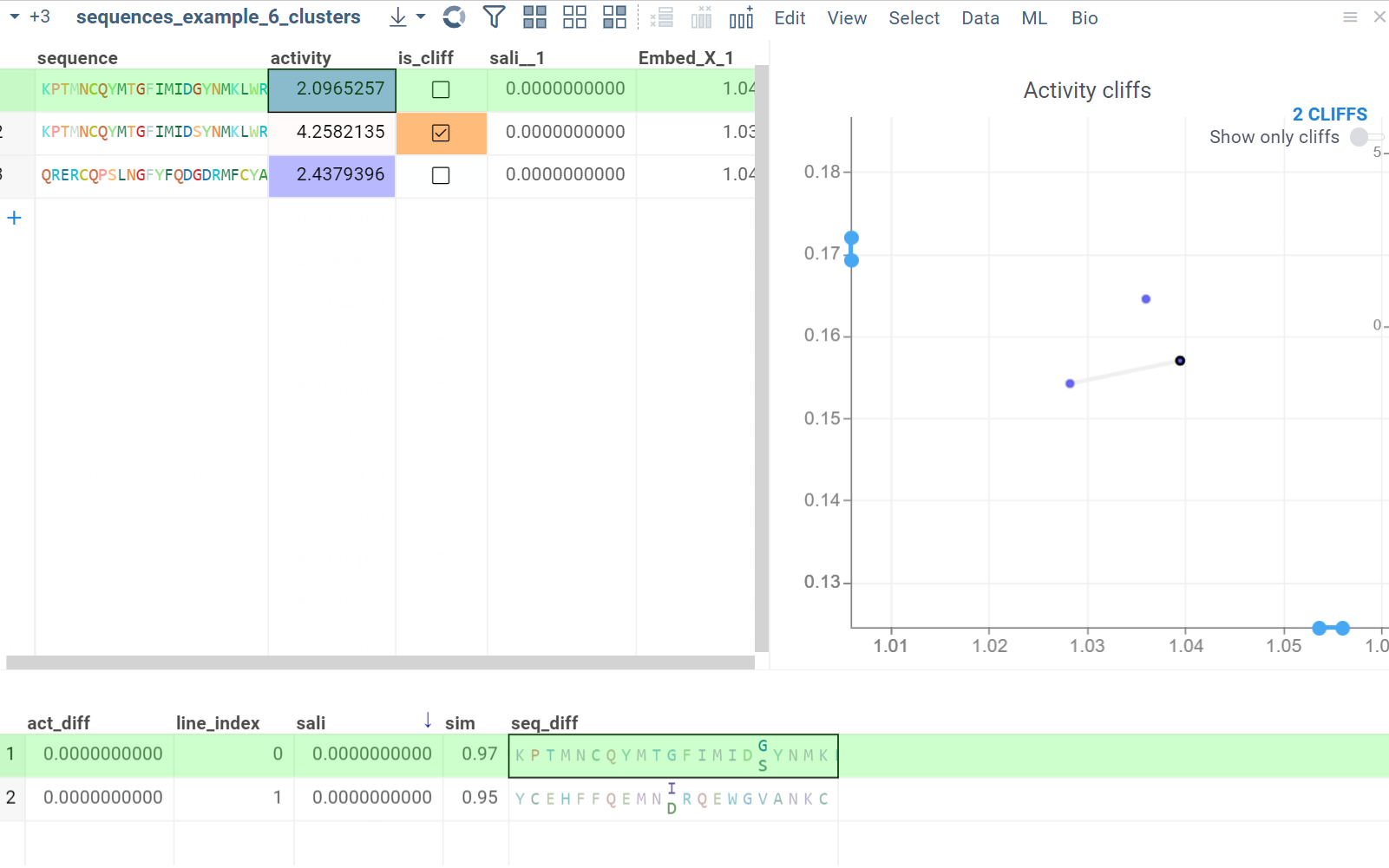
Activity cliffs
The Activity Cliffs tool in Datagrok detects and visualizes pairs of sequences with highly similar monomer composition but significantly different activity levels, known as "activity cliffs". The Activity Cliffs tool is an enhanced version of Sequence Space, specifically designed for Sequence-Activity Relationship (SAR) analysis. To run the analysis, you need a dataframe that contains peptide/DNA sequences along with numerical data representing the associated activity. For example, you can use sequences of short peptides with measured antimicrobial effects or DNA sequences with measured affinity to a specific protein.
- Activity Cliffs
- Cliff pairs
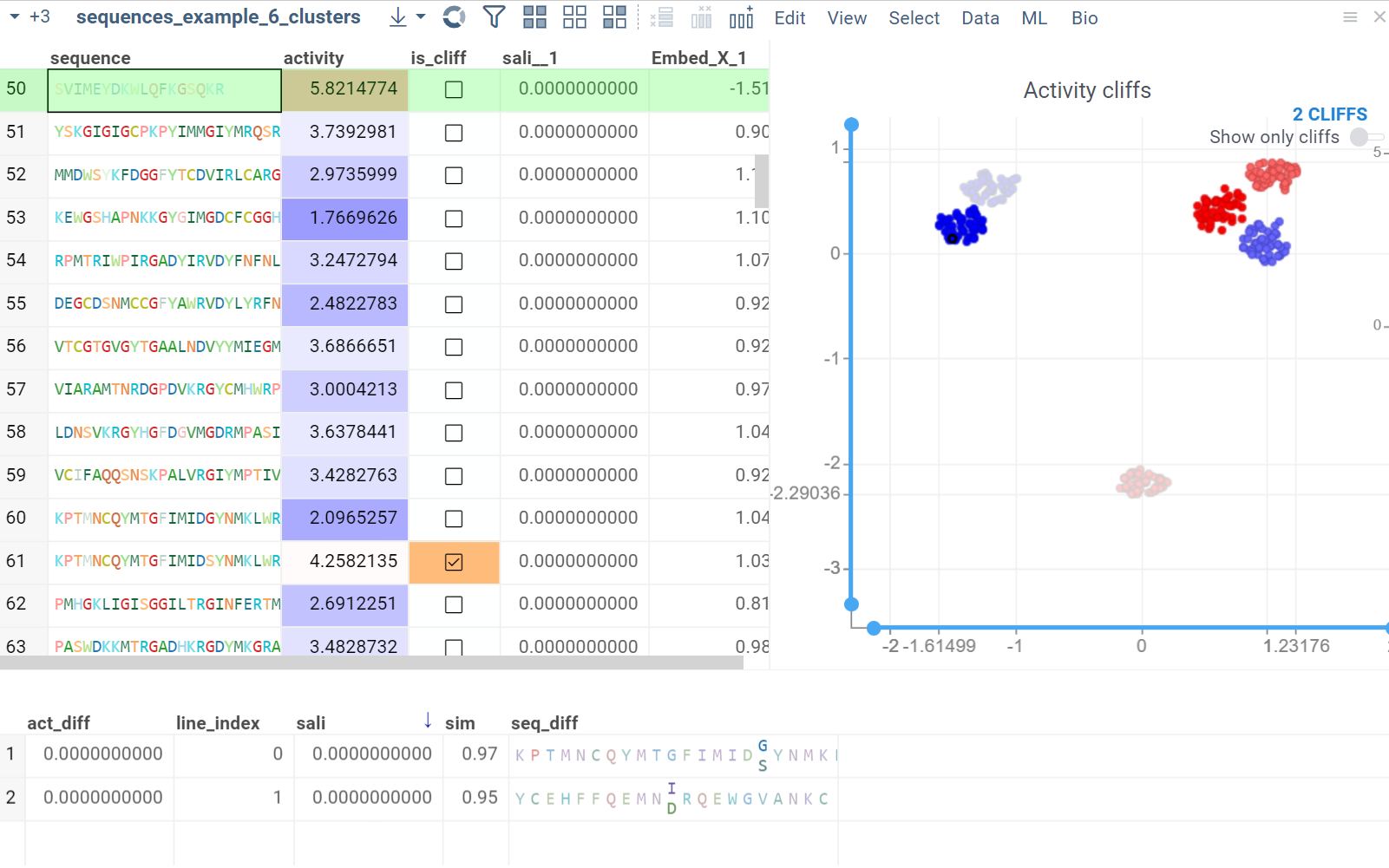
How to use
To run the activity cliffs analysis, do the following:
- In the Menu Ribbon, select Bio > Activity Cliffs... A parameter dialog opens.
- In the parameter dialog, specify the following:
- Select the source table, sequence column, and activity data column to analyze.
- Set the similarity cutoff.
- Select a dimensionality reduction algorithm and adjust its parameters using the Gear icon next to the Method control.
- Click OK to execute the analysis. A scatterplot visualization is added to the view.
- Optional. In the scatterplot, click the link with the detected number of cliffs to open an Activity Cliffs table containing all pairs of molecules identified as cliffs. The tables also has detailed information such as similarity score, activity difference score, and other data.
In the scatterplot, the marker color corresponds to the level of the sequence activity, and the size represents the maximum detected activity cliff for that sequence. The opacity of the line connecting sequence pairs corresponds to the size of the activity cliff.
To explore the sequence pairs:
- Click a sequence in the source dataframe to zoom in on the scatterplot and focus on the pair that includes the selected molecule. Hover over sequence pairs and connecting lines to see summary information about them.
- Click the line connecting sequences in the scatterplot to select a corresponding pair of sequences in the underlying dataframe and Activity Cliffs table. The reverse also applies: clicking a pair in the Activity Cliffs table updates the scatterplot and selects the corresponding rows in the underlying dataframe.
As you browse the dataset, the Context Panel updates with relevant information.
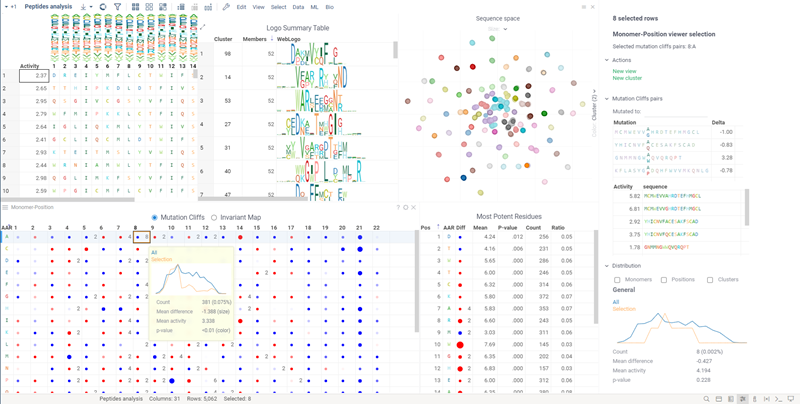
SAR for peptides
The Peptides application performs SAR analysis of peptides. The app offers the following features:
- Automatic detection of most potent monomer/positions.
- Filtering based on monomer, position, or any other attribute.
- Ability to analyze differences in activity distribution for groups of peptides.
- Dynamic calculations of statistically significant differences in activity distributions between groups.
- Analyzing the peptide space.
Utilities
Format conversion
Datagrok converts macromolecules between formats, such as HELM or FASTA.
For individual macromolecules, the conversion happens automatically as you interact with them in the dataset. The Context Panel shows all supported notations, along with the sequence view . You can also perform conversion on the entire column by choosing the corresponding option from the Bio > Convert menu.
Split to monomers
You can split linear macromolecules to monomers.
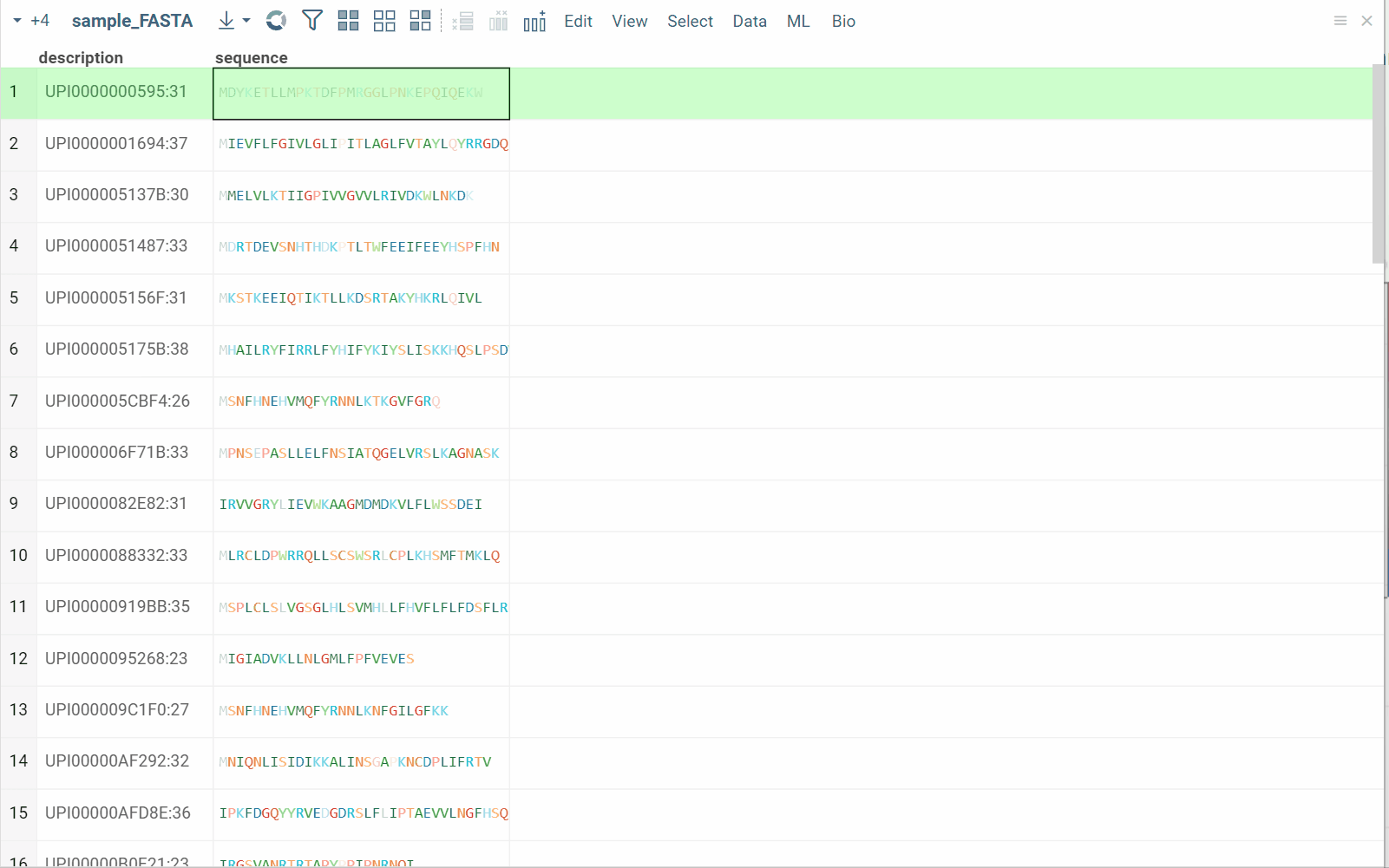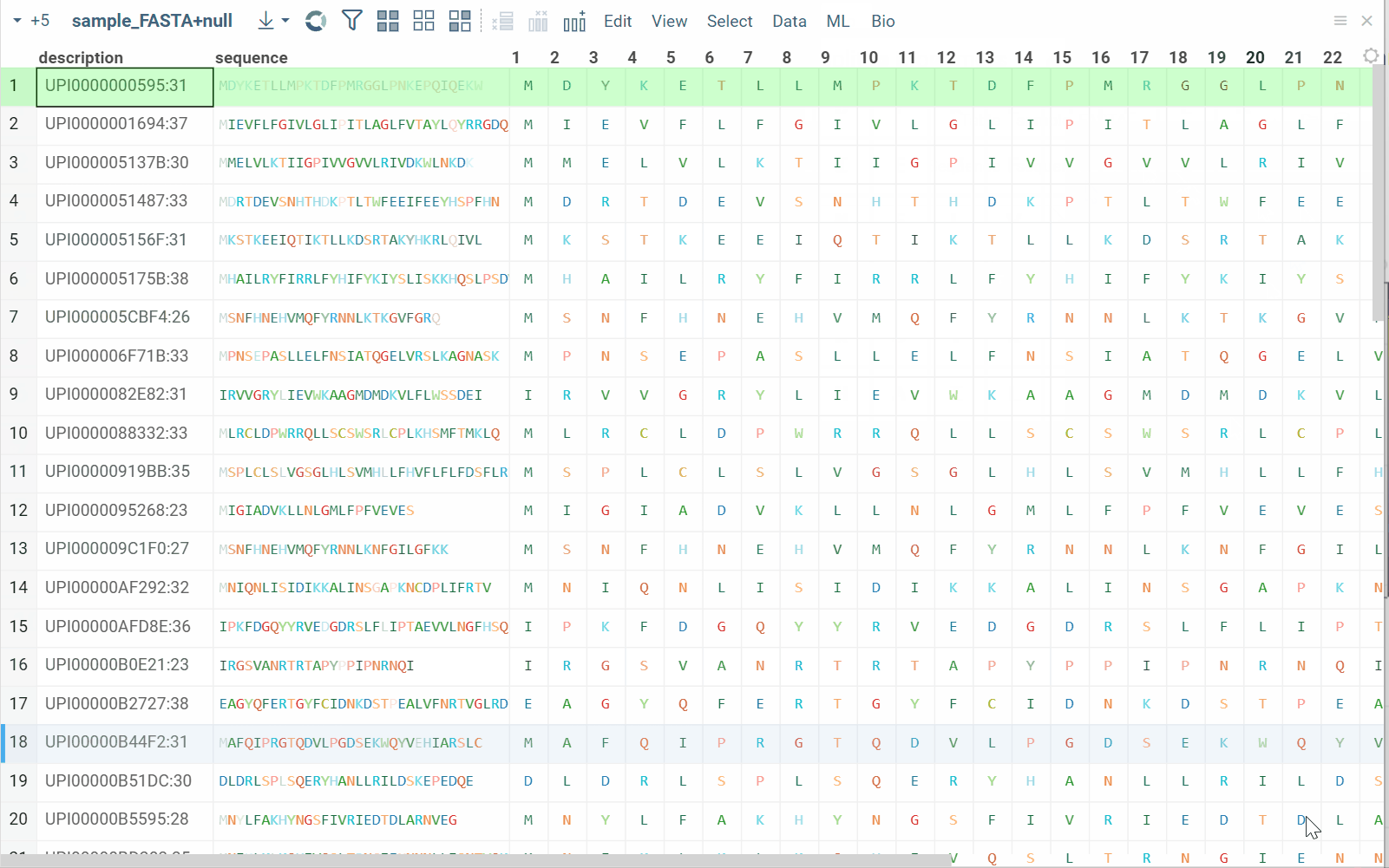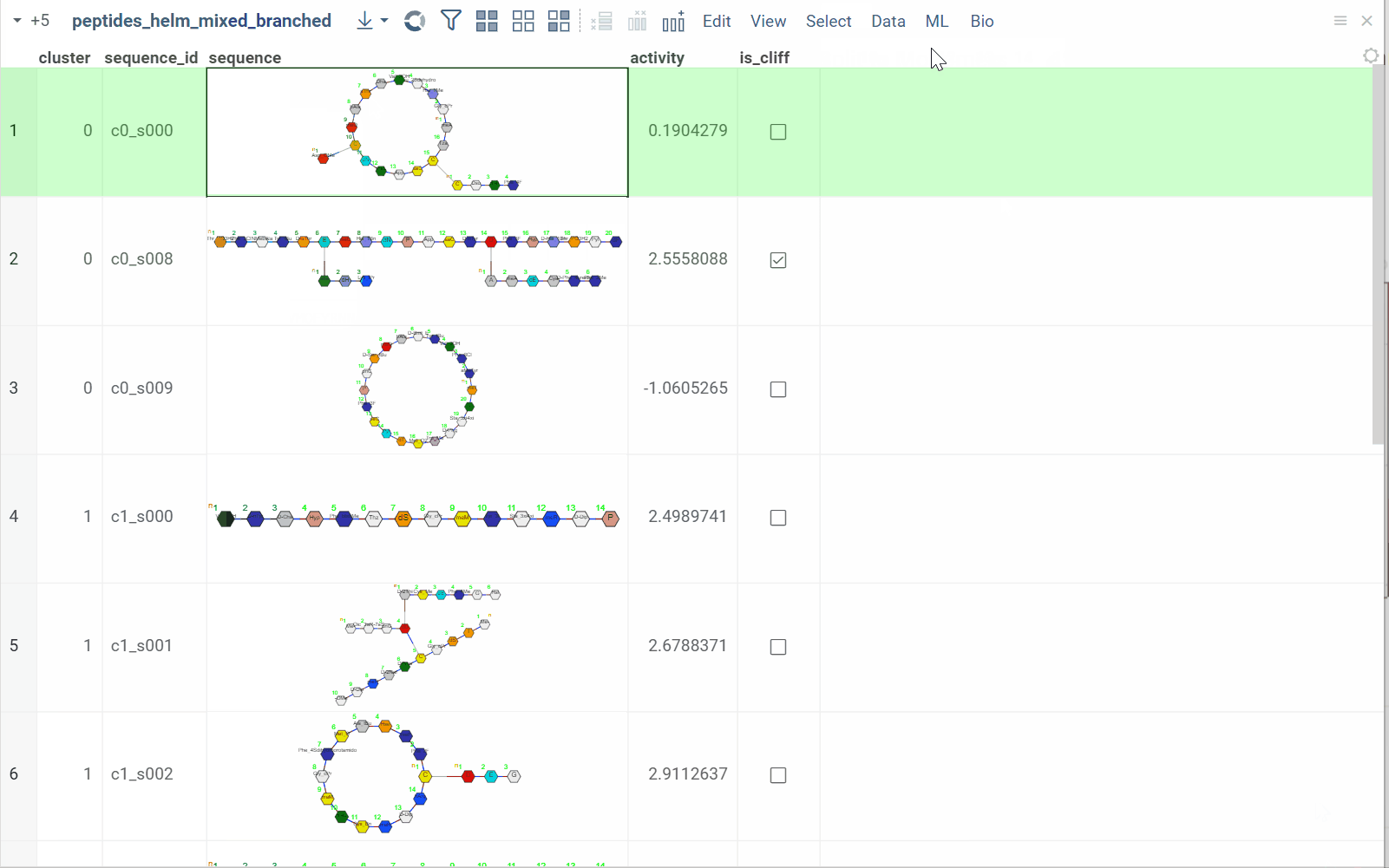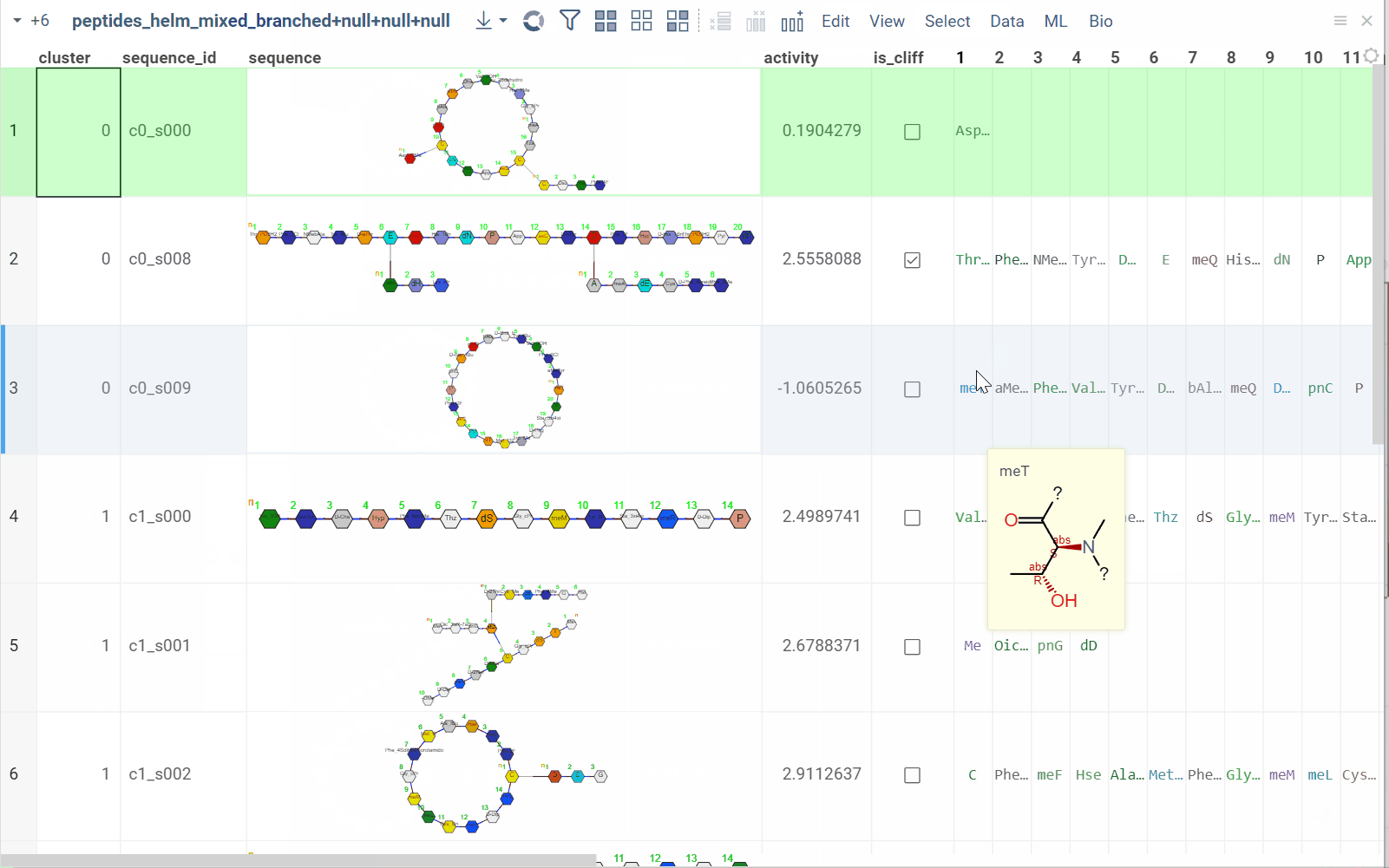
How to use
- In the Top Menu, select Bio > Convert > Split to Monomers. A dialog opens.
- In the dialog, select the sequence column and click OK to execute. New columns containing monomers are added to the table.
Get atomic-level structure
You have two options to generate the atomic structure of the sequences:
- Generate the sequence using the Helm Editor which results in the unordered molecule graph.
- For linear sequences, reproduce the linear form of molecules. This is useful for better visual inspection of a sequence and duplex comparison.
This approach could be used for any given case of HELM notation in order to get a visually appropriate form of monomers in cycles etc. Structure at atomic level could be saved in available notations.
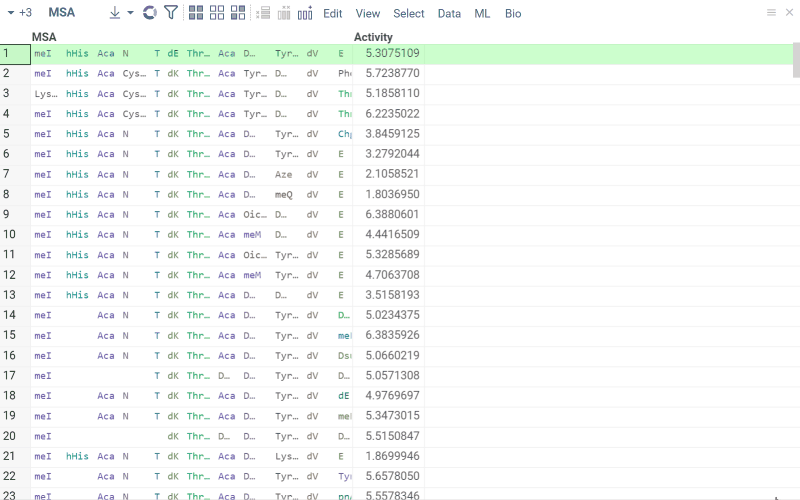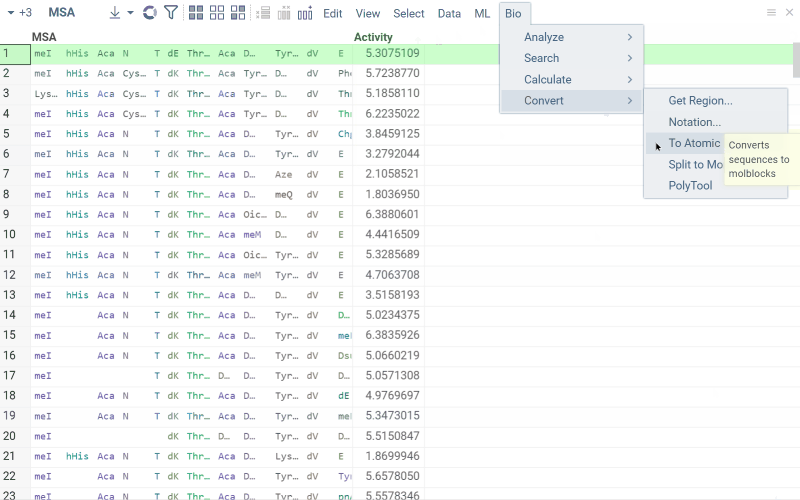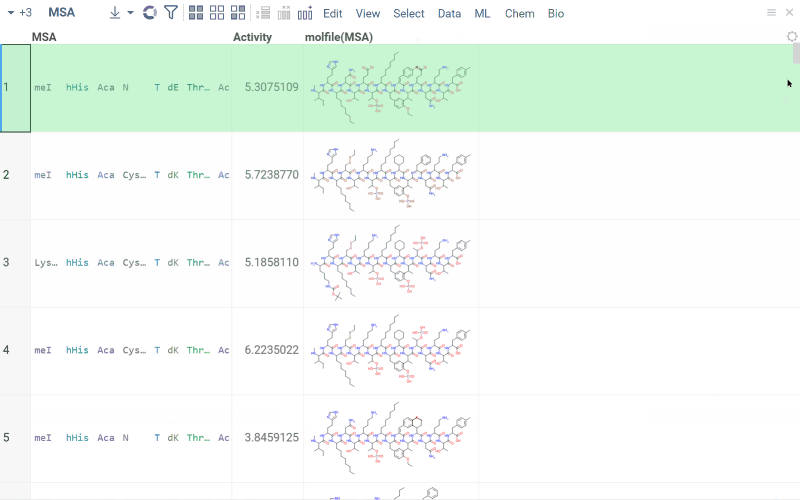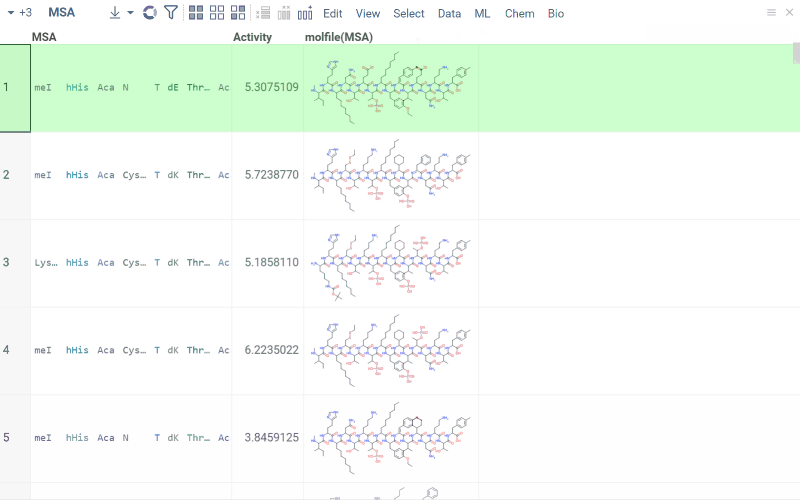
How to use
- In the Top Menu, select Bio > Convert > To Atomic Level. A dialog opens.
- In the dialog, select the sequence column and click OK to execute. A new column containing atomic structures of sequences is added to the table. In addition, a menu Chem appears in the Top Menu. Clicking the atomic-level structures displays cheminformatics-related information in the Context Panel.
Get region
With Datagrok, you can extract a region of sequences in a Macromolecule column.
The Get Region function maintains .positionNames and .positionLabels for extracted region.
The Get Region input form shows Region input if a Macromolecule column is annotated with .regions tag (JSON format),
to easy selection region of interest to extract.
How to use
-
To call Get Region:
- Select Bio > Convert > GetRegion. A dialog opens. In the dialog select a table and a sequence column.
- Click on the Hamburger icon of a Macromolecule column. Expand section Get Region.
-
Fill in start and end positions of the region of interest, and name of a column. A new column containing sequences of the region of interest is added to the table.
Oligo Toolkit
The Oligo Toolkit is a collection of tools helping you to work with oligonucleotide sequences. To learn more and how to use, see the OligoToolkit page.
Customizing and extending the platform
Datagrok is a highly flexible platform that can be tailored to meet your specific needs and requirements. With its comprehensive set of functions and scripting capabilities, you can customize and enhance any aspect of the platform to suit your biological data needs.
For instance, you can add new data formats, custom libraries, apply custom models, or perform advanced calculations and analyses using powerful bioinformatics libraries like Biopython, Bioconductor, ScanPy, and others.
You can also add or change UI elements, create custom connectors, menus, context actions, and more. You can even develop entire applications on top of the platform or customize any existing open-source plugins.
Learn more about extending and customizing Datagrok.