Use semantic types with scripts
The Datagrok semantic types is a very powerful concept, allowing you to define the meaning of your data. For example, you can specify that a particular string contains a chemical molecule in SMILES format, E-mail, or URL address.
For example, let's explore the Gasteiger partial charges script,
which takes a molecule in SMILES format as input
and calculates the Gasteiger charges distribution.
The script is provided with the Chem package.
The script takes as an input the string variable mol with the semantic annotation Molecule.
When you run the script, you will see the following:
- Result
- Python
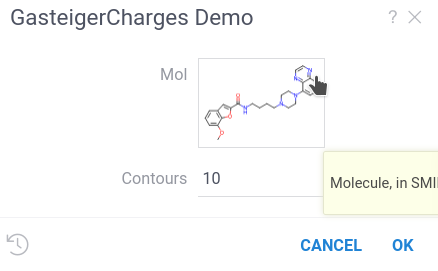
#name: GasteigerCharges Demo
#description: Calculates Gasteiger charge distribution
#language: python
#tags: demo, chem, rdkit
#input: string mol = "COc1cccc2cc(C(=O)NCCCCN3CCN(c4cccc5nccnc54)CC3)oc21" {semType: Molecule} [Molecule, in SMILES format]
#input: int contours = 10
#output: graphics charges [The Gasteiger partial charges]
from rdkit import Chem
from rdkit.Chem import AllChem
from rdkit.Chem.Draw import SimilarityMaps
mol = Chem.MolFromMolBlock(mol) if ("M END" in mol) else Chem.MolFromSmiles(mol)
if mol is not None:
AllChem.ComputeGasteigerCharges(mol)
contribs = [float(mol.GetAtomWithIdx(i).GetProp('_GasteigerCharge')) for i in range(mol.GetNumAtoms())]
charges = SimilarityMaps.GetSimilarityMapFromWeights(mol, contribs, contourLines=contours)
Datagrok recognized the Molecule semantic types and created the custom UI displaying molecule formula.
Click on it to open
chemical sketcher
and draw your own molecule.
Datagrok has outstanding chemoinformatics support,
so almost all UI elements provide you special viewing and editing options for chemical structures.
You can assign a semantic type to output variables in the same way. The semantic types annotation has no benefits for simple scalar output, but it is extremely helpful when you integrate your script with the Datagrok platform.
Semantic types for columns
Similarly, you can specify the semantic type for dataframe columns.
For example, let column or column_list selectors accept only
columns containing chemical molecules.
#input: dataframe df {caption: Dataframe}
#input: column mol {semType:Molecule; caption: Molecules} [Molecules to analyze]