NGL viewer
NGL viewer is used for visualizing and analyzing macromolecules.
It allows you to view 3D structures, explore docking results, and offers multiple rendering modes for different visual representations. The viewer is based on the NGL Viewer library.
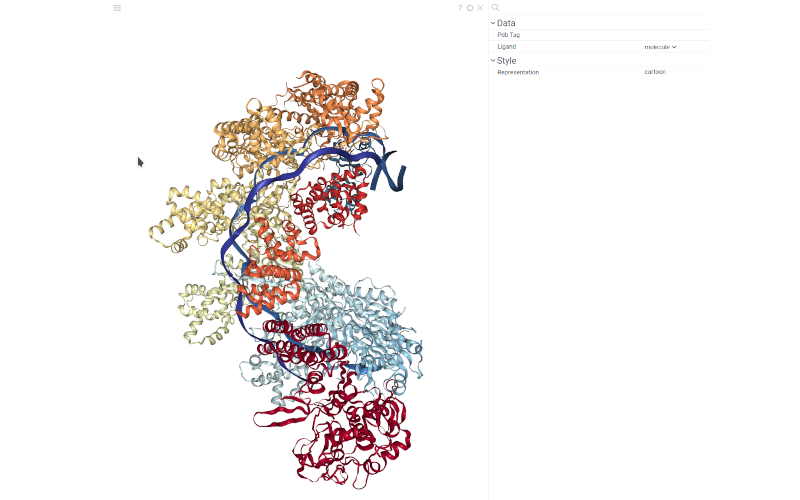
Creating an NGL viewer
- On the menu ribbon, click the Add viewer icon. A dialog opens.
- In the dialog, select NGL.
- Load the macromolecule file by using the "Open..." link in the center of the viewer.
Note: The user interface for loading macromolecules from a file share is not implemented. You have to save the macromolecule to your local device before opening it in the NGL Viewer.
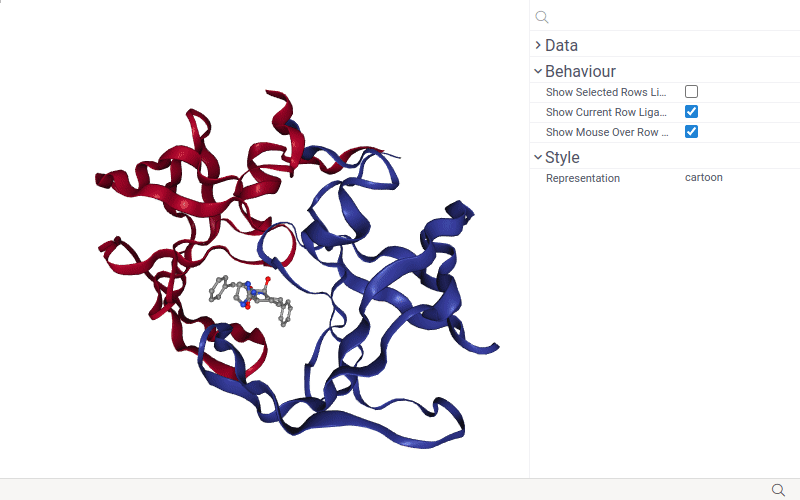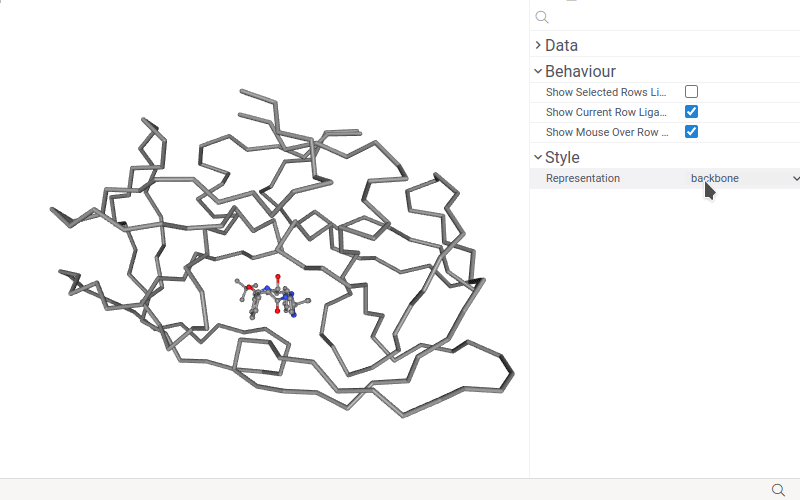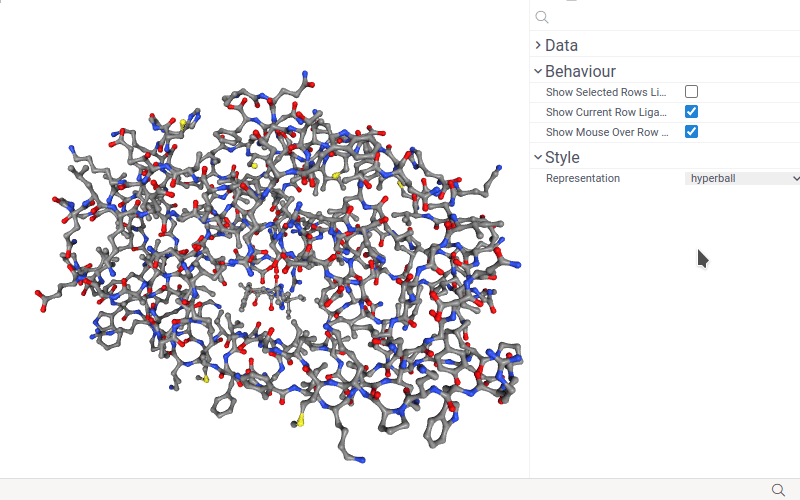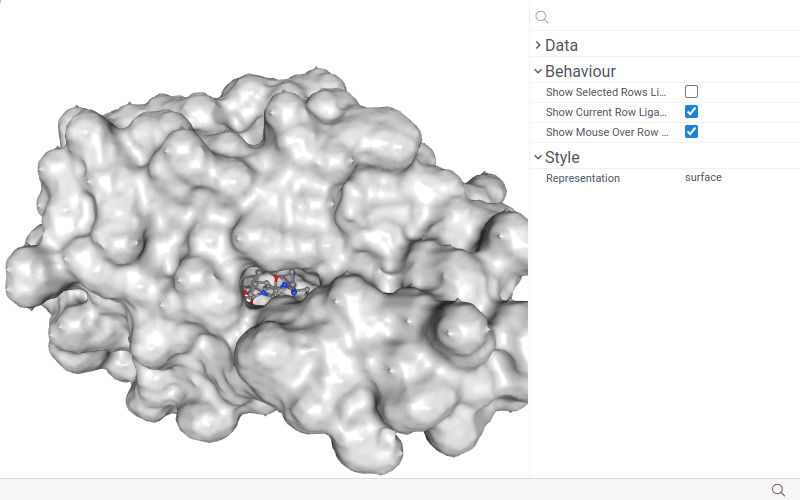
The NGL viewer detects a column of semantic type 'Molecule' in the table and displays molecules along with the structure.
Configuring an NGL viewer
You can set the source column for the small molecules and customize visualization options. To do that, click the Gear icon on top of the viewer and use the Data and Style info panes on the Context Panel.
You can:
- Select the ligand column using the Ligand control.
- Change macromolecule representation mode using the Representation control.
Interaction with other viewers
The NGL viewer shows small molecules (ligands) together with the macromolecule when you select it or just hover mouse over it.
If only one ligand is selected, the ligand is displayed with a full-color ball+stick representation. If there are multiple selected ligands, then the ligand of the current row is displayed in 'green', the mouse-over row ligand is displayed in 'light gray', and selected rows ligands are displayed in orange.
Viewer controls
| Action | Control |
|---|---|
| View info for the specific atom | Hover mouse over the atom |
| Move the atom to the viewer center | Click the atom |