Position Statistics
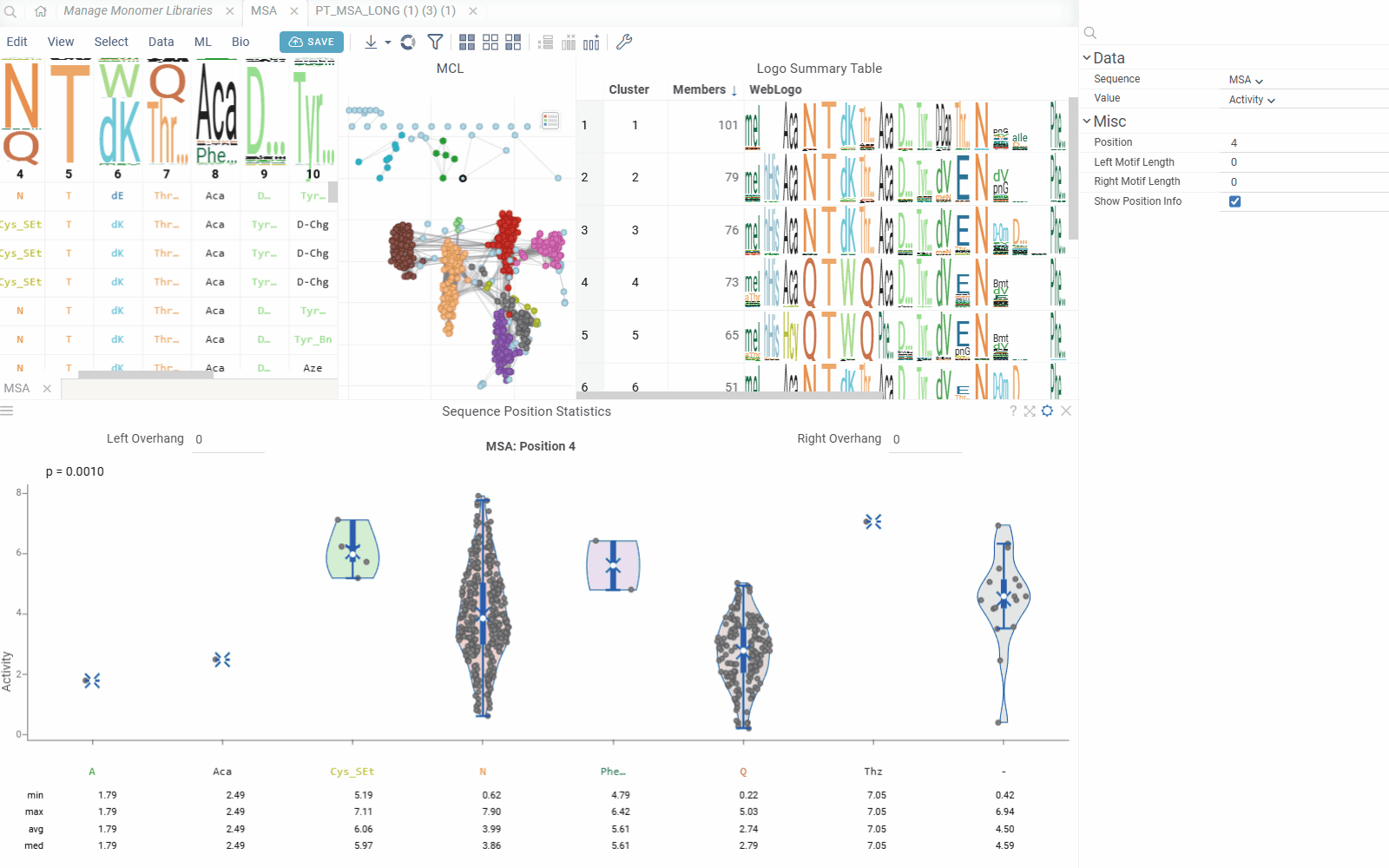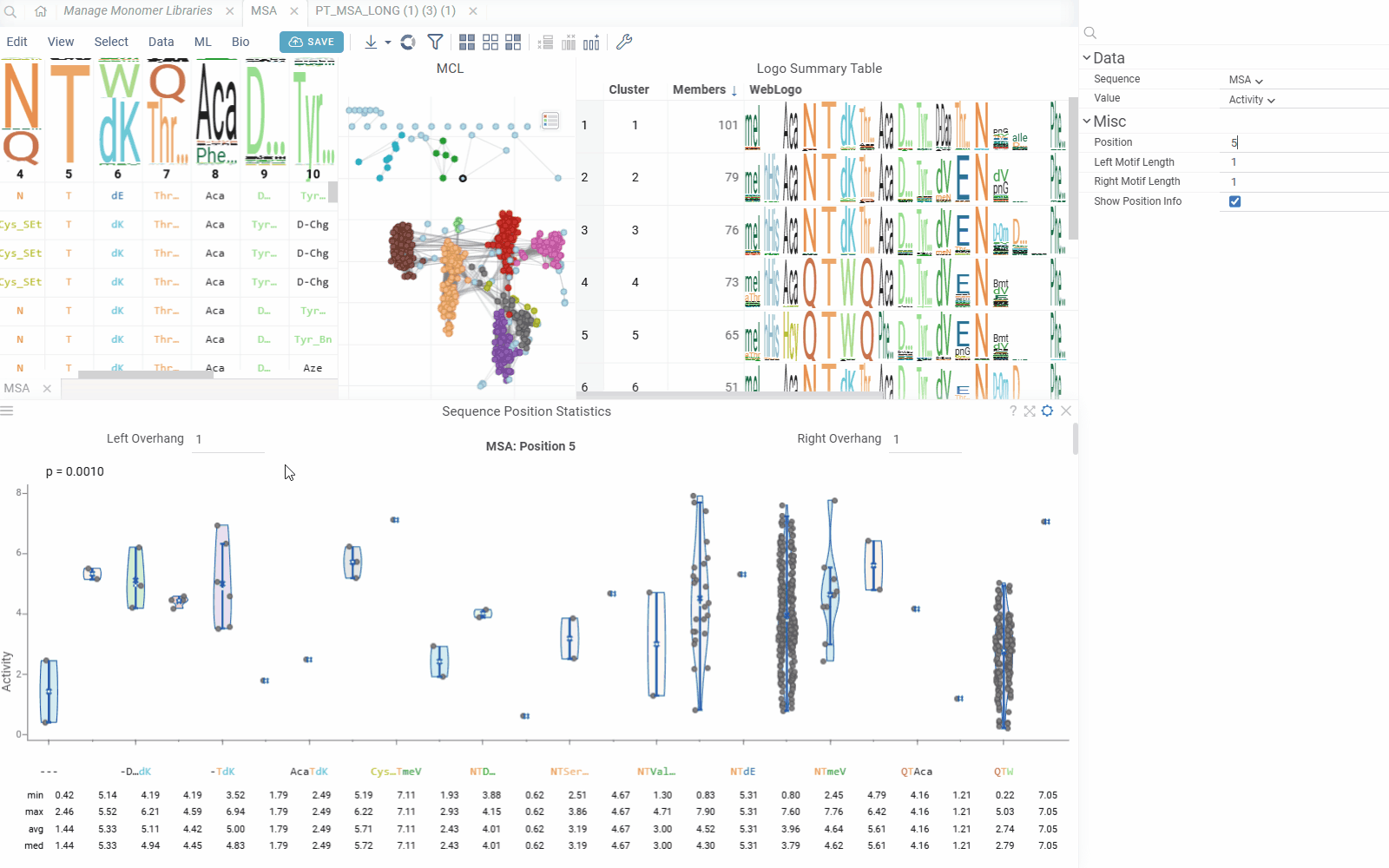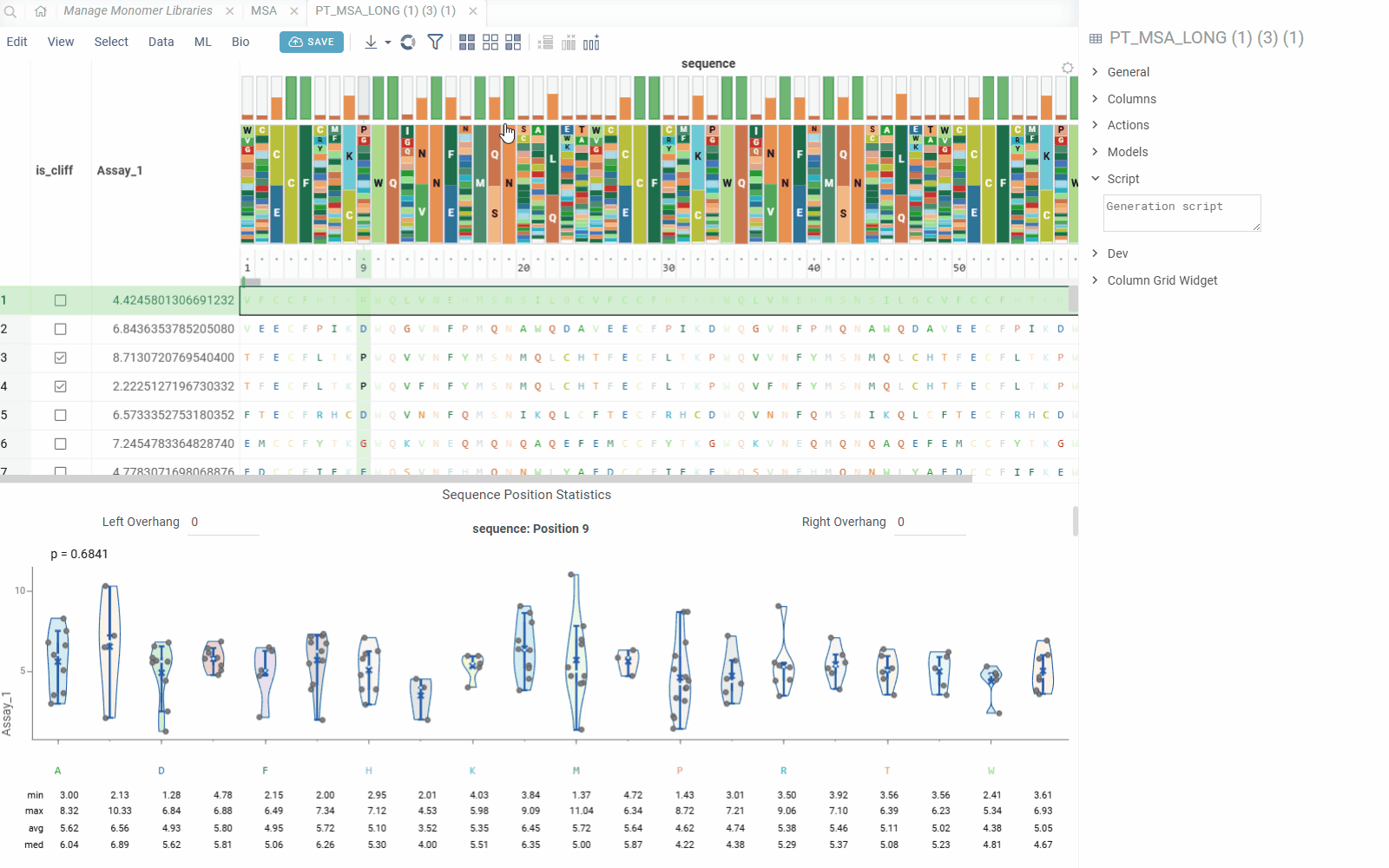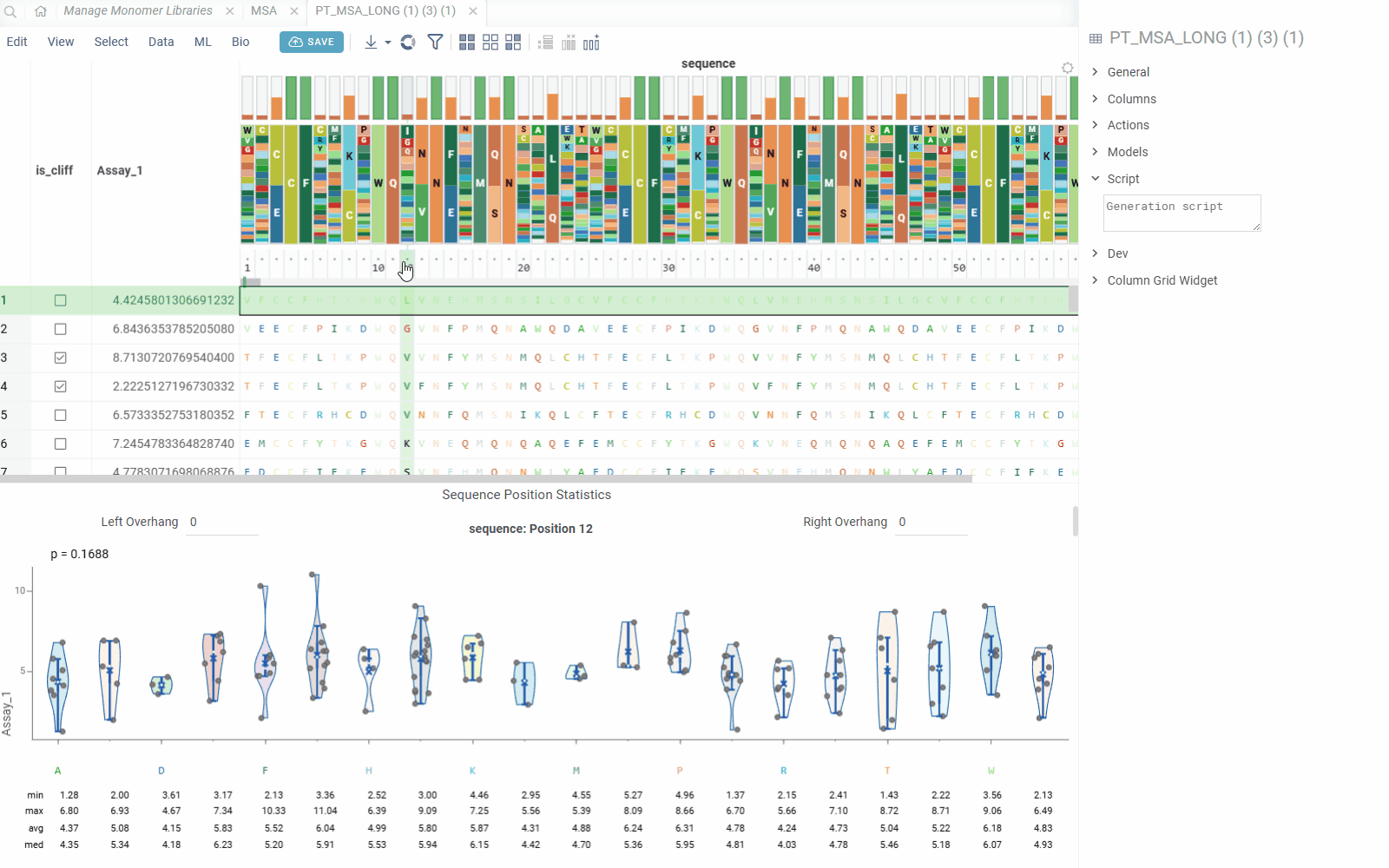
The Position Statistics viewer displays statistical distributions of values at specific sequence positions using violin/box plots. This viewer helps analyze how values vary across different monomers at a selected position, with optional motif context visualization.
Features
- Position Selection: Choose any position in the sequence for statistical analysis
- Motif Context: Configure left and right overhang lengths to include neighboring positions
- Value Distribution: Visualize value distributions using violin plots with statistical summaries
- Interactive Navigation: Easy position switching with automatic updates
- Configurable Display: Option to show/hide position and other statistical information in the viewer header and footer.
Customization
You can modify the Position Statistics Viewer through the Context Panel:
- Sequence: The column containing sequence data in any notation
- Value: The column containing numeric values for statistical analysis
- Position: The position in the sequence to analyze (1-based indexing)
- Left Motif Length: Number of positions to include left of the selected position (0-10)
- Right Motif Length: Number of positions to include right of the selected position (0-10)
- Show Position Info: Toggle display of position and overhang controls in the header
Data requirements
The viewer requires:
- A sequence column with macromolecule data (FASTA, HELM, or separated format)
- A numeric column with values for statistical analysis
Position context
The viewer creates motif representations by combining monomers from multiple positions:
- Center Position: The primary position being analyzed
- Left Overhang: Positions to the left of the center (configurable 0-10)
- Right Overhang: Positions to the right of the center (configurable 0-10)
Monomers from all positions in the motif are joined to create unique combinations for statistical analysis.
Statistical display
The viewer shows statistics for each monomer(s) combination:
- Min/Max: Minimum and maximum values
- Average: Mean value across sequences
- Median: Median value
- Count: Number of sequences with each monomer combination
- Standard Deviation: Measure of value dispersion
Interactivity
The Position Statistics Viewer follows standard Datagrok patterns:
- Position Controls: Use the header controls to adjust left/right overhang lengths
- Value Column Selection: The viewer automatically updates when you change the value column
- Responsive Layout: Automatically adjusts to show/hide position information based on settings
Integration with Peptides SAR analysis
When used within Peptides SAR analysis, the Position Statistics Viewer automatically:
- Synchronizes with the selected position from other viewers
- Uses the same sequence column as the main analysis
- Updates position selection based on sequence column metadata
For standalone use, the viewer generates position columns automatically from the sequence data.
Position selection
The viewer can automatically detect the current position from sequence column metadata. When integrated with other sequence analysis tools, position changes are synchronized across viewers for consistent analysis.