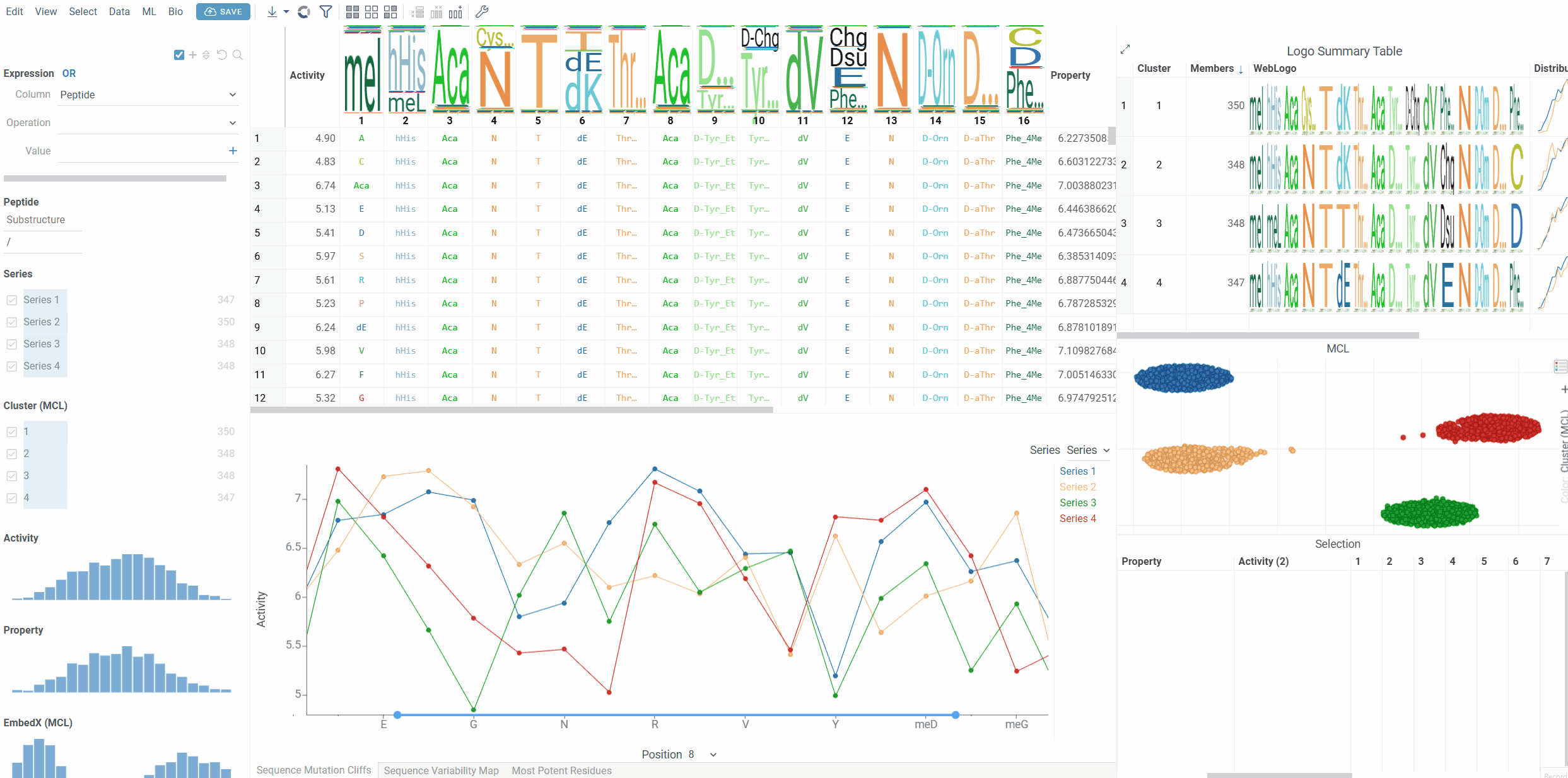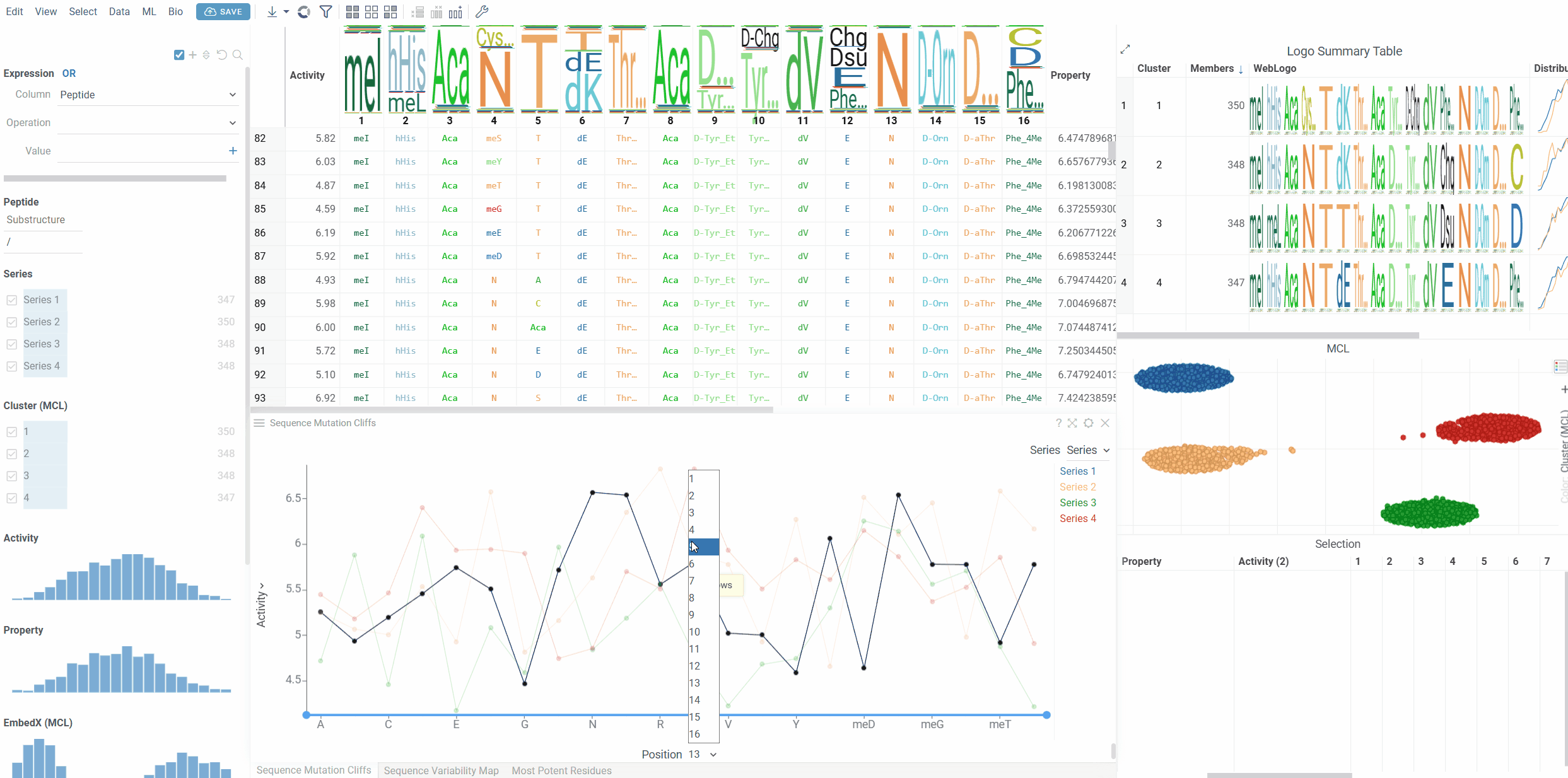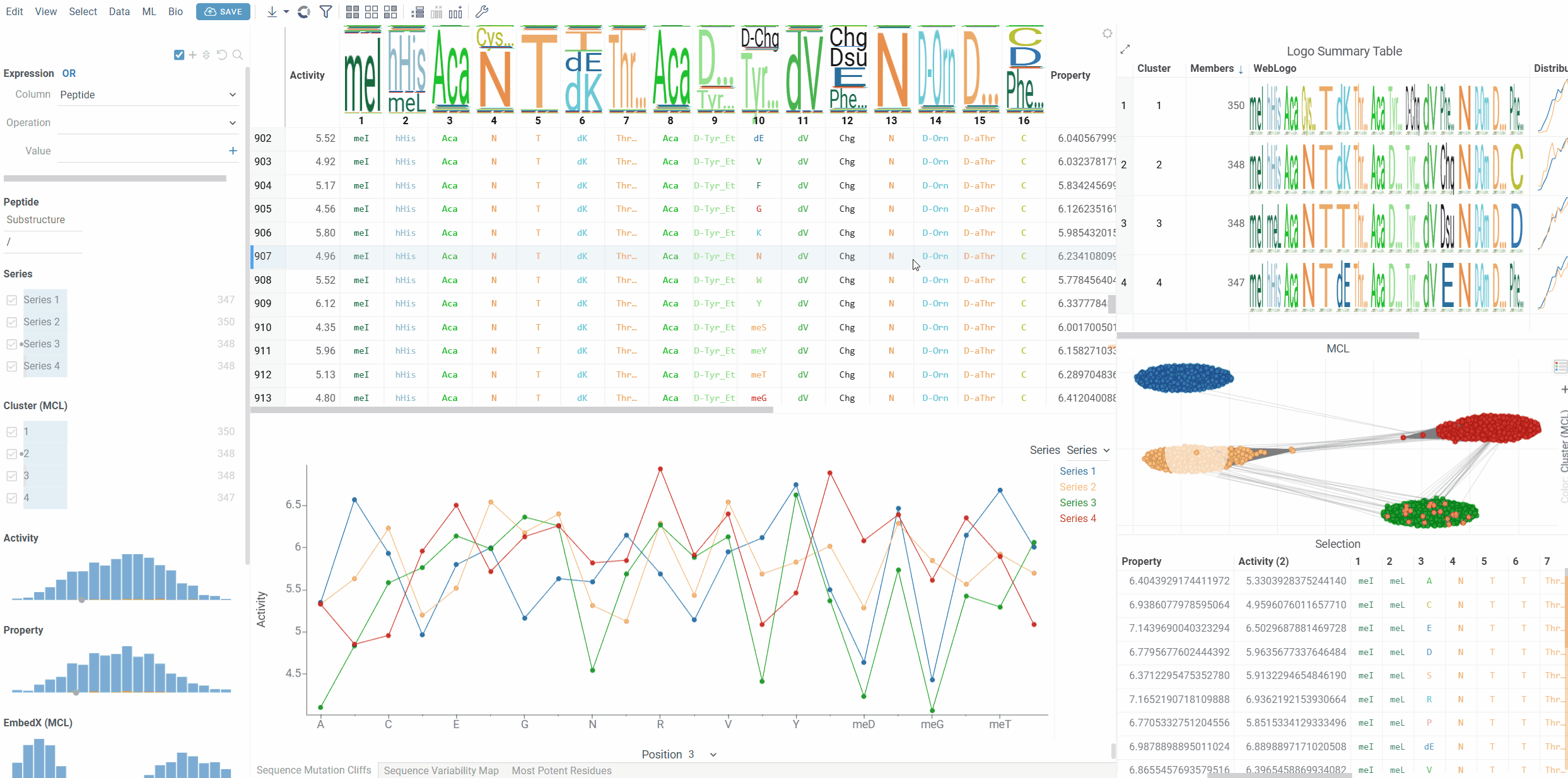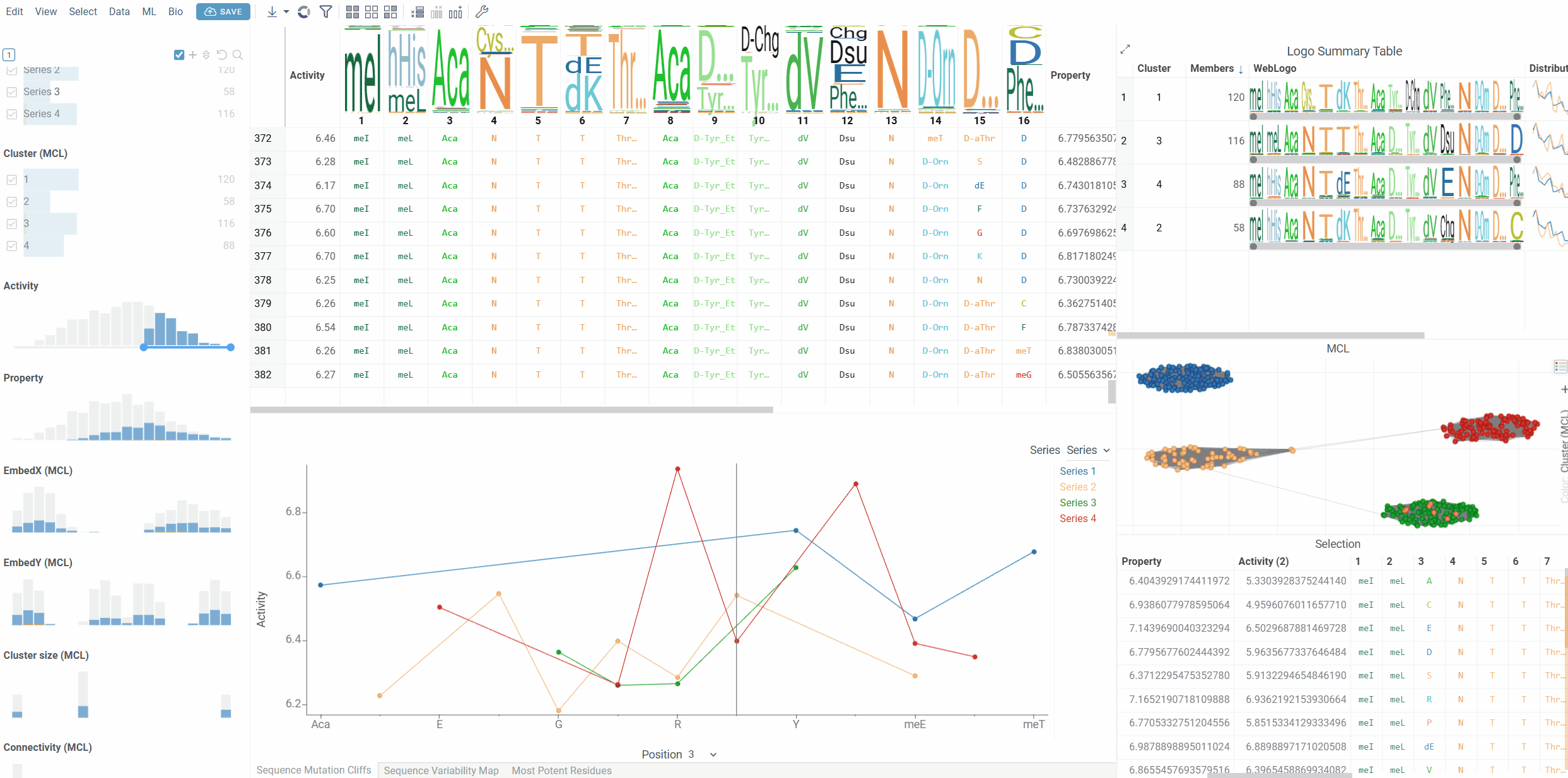
Mutation Cliffs Viewer
The Mutation Cliffs Viewer displays sequence mutation cliffs as a line chart, showing how single amino acid changes at specific positions affect activity values. This viewer helps identify critical positions where small structural changes lead to significant activity differences.
The viewer shows mutation cliff pairs at a selected position, with each point representing a sequence that participates in a mutation cliff. When series data is available, different series are displayed as separate lines with distinct colors.
Features
- Position Analysis: Select any position in the sequence to analyze mutation cliffs at that specific location
- Series Grouping: Group mutation cliffs by categorical variables (e.g., experimental series, compound classes)
- Interactive Selection: Click points to select corresponding sequences in the main dataset
- Current Row Following: Option to show only mutation cliffs related to the currently selected sequence
- Logarithmic Scaling: Switch between linear and logarithmic Y-axis scaling
Customization
You can modify the Mutation Cliffs Viewer through the property panel:
- Sequence: The column containing sequence data (macromolecule format)
- Activity: The column containing numeric activity data
- Series: Optional categorical column for grouping sequences into series
- Position: The position in the sequence to analyze (1-based indexing)
- Y-Axis Type: Choose between Linear or Logarithmic scaling
- Current Row Mutations Only: When enabled, shows only mutation cliffs involving the currently selected sequence
Data Requirements
The viewer requires:
- A sequence column with macromolecule data (FASTA, HELM, BILN or separated format)
- An activity column with numeric values
- Sequences must have mutation cliffs at the selected position (sequences differing by exactly one amino acid with some activity differences). If not, the viewer will display no data points.
Interactivity
The Mutation Cliffs Viewer follows standard Datagrok selection patterns:
- Click: Select individual sequences
- Ctrl+Click: Toggle sequence selection
- Shift+Click: Add sequences to selection
- Hover: Highlight corresponding sequences across all viewers
The viewer maintains bidirectional selection synchronization with the main dataset - selecting points in the line chart selects the corresponding rows in the main table, and vice versa.
Position Navigation
Use the position selector at the bottom of the viewer to switch between different sequence positions. Only positions that contain mutation cliffs will show data points.
Integration with SAR Analysis
When used within Peptides SAR analysis, the Mutation Cliffs Viewer automatically:
- Uses position columns generated during SAR analysis
- Inherits mutation cliffs calculations from other viewers
- Synchronizes with other SAR viewers for consistent selection and highlighting
For datasets without existing SAR analysis, the viewer generates position columns automatically from the sequence data.